Tyrosyl-DNA phosphodiesterase I catalytic mutants reveal an alternative nucleophile that can catalyze substrate cleavage
- PMID: 25609251
- PMCID: PMC4358259
- DOI: 10.1074/jbc.M114.635284
Tyrosyl-DNA phosphodiesterase I catalytic mutants reveal an alternative nucleophile that can catalyze substrate cleavage
Abstract
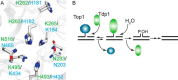
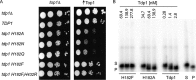
Tyrosyl-DNA phosphodiesterase I (Tdp1) catalyzes the repair of 3'-DNA adducts, such as the 3'-phosphotyrosyl linkage of DNA topoisomerase I to DNA. Tdp1 contains two conserved catalytic histidines: a nucleophilic His (His(nuc)) that attacks DNA adducts to form a covalent 3'-phosphohistidyl intermediate and a general acid/base His (His(gab)), which resolves the Tdp1-DNA linkage. A His(nuc) to Ala mutant protein is reportedly inactive, whereas the autosomal recessive neurodegenerative disease SCAN1 has been attributed to the enhanced stability of the Tdp1-DNA intermediate induced by mutation of His(gab) to Arg. However, here we report that expression of the yeast His(nuc)Ala (H182A) mutant actually induced topoisomerase I-dependent cytotoxicity and further enhanced the cytotoxicity of Tdp1 His(gab) mutants, including H432N and the SCAN1-related H432R. Moreover, the His(nuc)Ala mutant was catalytically active in vitro, albeit at levels 85-fold less than that observed with wild type Tdp1. In contrast, the His(nuc)Phe mutant was catalytically inactive and suppressed His(gab) mutant-induced toxicity. These data suggest that the activity of another nucleophile when His(nuc) is replaced with residues containing a small side chain (Ala, Asn, and Gln), but not with a bulky side chain. Indeed, genetic, biochemical, and mass spectrometry analyses show that a highly conserved His, immediately N-terminal to His(nuc), can act as a nucleophile to catalyze the formation of a covalent Tdp1-DNA intermediate. These findings suggest that the flexibility of Tdp1 active site residues may impair the resolution of mutant Tdp1 covalent phosphohistidyl intermediates and provide the rationale for developing chemotherapeutics that stabilize the covalent Tdp1-DNA intermediate.
Keywords: DNA Damage; DNA Repair; DNA Topoisomerase; Enzyme Mechanism; Enzyme Mutation; Protein-DNA Covalent Complexes; Spinocerebellar Ataxia with Axonal Neuropathy 1; Tyrosyl-DNA Phosphodiesterase I.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target.Drug Metab Rev. 2014 Nov;46(4):494-507. doi: 10.3109/03602532.2014.971957. Epub 2014 Oct 20. Drug Metab Rev. 2014. PMID: 25327705 Review.
-
Analysis of the active-site mechanism of tyrosyl-DNA phosphodiesterase I: a member of the phospholipase D superfamily.J Mol Biol. 2012 Jan 27;415(4):741-58. doi: 10.1016/j.jmb.2011.11.044. Epub 2011 Dec 6. J Mol Biol. 2012. PMID: 22155078 Free PMC article.
-
Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison.J Mol Biol. 2007 Sep 28;372(4):1070-1081. doi: 10.1016/j.jmb.2007.07.055. Epub 2007 Aug 2. J Mol Biol. 2007. PMID: 17707402
-
In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation.DNA Repair (Amst). 2009 May 1;8(5):654-63. doi: 10.1016/j.dnarep.2008.12.012. Epub 2009 Feb 10. DNA Repair (Amst). 2009. PMID: 19211312 Free PMC article.
-
Spinocerebellar ataxia with axonal neuropathy.Adv Exp Med Biol. 2010;685:75-83. doi: 10.1007/978-1-4419-6448-9_7. Adv Exp Med Biol. 2010. PMID: 20687496 Review.
Cited by
-
N-terminal domain of tyrosyl-DNA phosphodiesterase I regulates topoisomerase I-induced toxicity in cells.Sci Rep. 2023 Jan 25;13(1):1377. doi: 10.1038/s41598-023-28564-6. Sci Rep. 2023. PMID: 36697463 Free PMC article.
-
Tyrosyl-DNA Phosphodiesterase I N-Terminal Domain Modifications and Interactions Regulate Cellular Function.Genes (Basel). 2019 Nov 6;10(11):897. doi: 10.3390/genes10110897. Genes (Basel). 2019. PMID: 31698852 Free PMC article. Review.
-
Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.Comput Struct Biotechnol J. 2021 May 25;19:3293-3302. doi: 10.1016/j.csbj.2021.05.035. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34188778 Free PMC article.
-
Topoisomerases and cancer chemotherapy: recent advances and unanswered questions.F1000Res. 2019 Sep 30;8:F1000 Faculty Rev-1704. doi: 10.12688/f1000research.20201.1. eCollection 2019. F1000Res. 2019. PMID: 31602296 Free PMC article. Review.
-
Abacavir, an anti-HIV-1 drug, targets TDP1-deficient adult T cell leukemia.Sci Adv. 2015 Apr 24;1(3):e1400203. doi: 10.1126/sciadv.1400203. eCollection 2015 Apr. Sci Adv. 2015. PMID: 26601161 Free PMC article.
References
-
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260, 14873–14878 - PubMed
-
- Hsiang Y. H., Lihou M. G., Liu L. F. (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49, 5077–5082 - PubMed
-
- Schoeffler A. J., Berger J. M. (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

