Transgenic plants as low-cost platform for chemotherapeutic drugs screening
- PMID: 25608652
- PMCID: PMC4307356
- DOI: 10.3390/ijms16012174
Transgenic plants as low-cost platform for chemotherapeutic drugs screening
Abstract
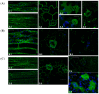
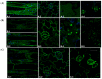
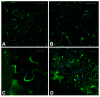
In this work we explored the possibility of using genetically modified Arabidopsis thaliana plants as a rapid and low-cost screening tool for evaluating human anticancer drugs action and efficacy. Here, four different inhibitors with a validated anticancer effect in humans and distinct mechanism of action were screened in the plant model for their ability to interfere with the cytoskeletal and endomembrane networks. We used plants expressing a green fluorescent protein (GFP) tagged microtubule-protein (TUA6-GFP), and three soluble GFPs differently sorted to reside in the endoplasmic reticulum (GFPKDEL) or to accumulate in the vacuole through a COPII dependent (AleuGFP) or independent (GFPChi) mechanism. Our results demonstrated that drugs tested alone or in combination differentially influenced the monitored cellular processes including cytoskeletal organization and endomembrane trafficking. In conclusion, we demonstrated that A. thaliana plants are sensitive to the action of human chemotherapeutics and can be used for preliminary screening of drugs efficacy. The cost-effective subcellular imaging in plant cell may contribute to better clarify drugs subcellular targets and their anticancer effects.
Figures
Similar articles
-
Cisplatin, Oxaliplatin, and Kiteplatin Subcellular Effects Compared in a Plant Model.Int J Mol Sci. 2017 Jan 31;18(2):306. doi: 10.3390/ijms18020306. Int J Mol Sci. 2017. PMID: 28146116 Free PMC article.
-
Investigation of Drug Efficacy by Screening Bioactive Chemical Effects on Plant Cell Subcellular Architecture.Methods Mol Biol. 2021;2213:49-58. doi: 10.1007/978-1-0716-0954-5_5. Methods Mol Biol. 2021. PMID: 33270192
-
Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles.Plant Cell Physiol. 2014 Apr;55(4):715-26. doi: 10.1093/pcp/pcu041. Epub 2014 Feb 23. Plant Cell Physiol. 2014. PMID: 24566535
-
Towards a better recording of microtubule cytoskeletal spatial organization and dynamics in plant cells.J Integr Plant Biol. 2019 Apr;61(4):388-393. doi: 10.1111/jipb.12721. Epub 2018 Nov 26. J Integr Plant Biol. 2019. PMID: 30226291
-
Selective chemical probes can untangle the complexity of the plant cell endomembrane system.Curr Opin Plant Biol. 2022 Aug;68:102223. doi: 10.1016/j.pbi.2022.102223. Epub 2022 May 11. Curr Opin Plant Biol. 2022. PMID: 35567926 Review.
Cited by
-
In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators.Int J Mol Sci. 2018 Jul 23;19(7):2135. doi: 10.3390/ijms19072135. Int J Mol Sci. 2018. PMID: 30041423 Free PMC article.
-
A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.iScience. 2023 Sep 20;26(10):107919. doi: 10.1016/j.isci.2023.107919. eCollection 2023 Oct 20. iScience. 2023. PMID: 37822503 Free PMC article.
-
Cisplatin, Oxaliplatin, and Kiteplatin Subcellular Effects Compared in a Plant Model.Int J Mol Sci. 2017 Jan 31;18(2):306. doi: 10.3390/ijms18020306. Int J Mol Sci. 2017. PMID: 28146116 Free PMC article.
-
Glutathione S-transferase related detoxification processes are correlated with receptor-mediated vacuolar sorting mechanisms.Plant Cell Rep. 2017 Sep;36(9):1361-1373. doi: 10.1007/s00299-017-2159-3. Epub 2017 Jun 2. Plant Cell Rep. 2017. PMID: 28577236
-
Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation.Int J Mol Sci. 2020 Jul 12;21(14):4925. doi: 10.3390/ijms21144925. Int J Mol Sci. 2020. PMID: 32664691 Free PMC article. Review.
References
-
- Orlikova B., Legrand N., Panning J., Dicato M., Diederich M. Anti-inflammatory and anticancer drugs from nature. Cancer Treat. Res. 2014;159:123–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

