Analytical study of robustness of a negative feedback oscillator by multiparameter sensitivity
- PMID: 25605374
- PMCID: PMC4305980
- DOI: 10.1186/1752-0509-8-S5-S1
Analytical study of robustness of a negative feedback oscillator by multiparameter sensitivity
Abstract
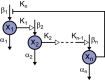
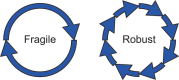
Background: One of the distinctive features of biological oscillators such as circadian clocks and cell cycles is robustness which is the ability to resume reliable operation in the face of different types of perturbations. In the previous study, we proposed multiparameter sensitivity (MPS) as an intelligible measure for robustness to fluctuations in kinetic parameters. Analytical solutions directly connect the mechanisms and kinetic parameters to dynamic properties such as period, amplitude and their associated MPSs. Although negative feedback loops are known as common structures to biological oscillators, the analytical solutions have not been presented for a general model of negative feedback oscillators.
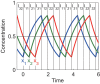
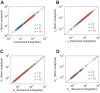
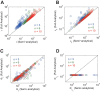
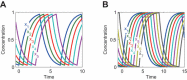
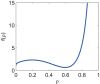
Results: We present the analytical expressions for the period, amplitude and their associated MPSs for a general model of negative feedback oscillators. The analytical solutions are validated by comparing them with numerical solutions. The analytical solutions explicitly show how the dynamic properties depend on the kinetic parameters. The ratio of a threshold to the amplitude has a strong impact on the period MPS. As the ratio approaches to one, the MPS increases, indicating that the period becomes more sensitive to changes in kinetic parameters. We present the first mathematical proof that the distributed time-delay mechanism contributes to making the oscillation period robust to parameter fluctuations. The MPS decreases with an increase in the feedback loop length (i.e., the number of molecular species constituting the feedback loop).
Conclusions: Since a general model of negative feedback oscillators was employed, the results shown in this paper are expected to be true for many of biological oscillators. This study strongly supports that the hypothesis that phosphorylations of clock proteins contribute to the robustness of circadian rhythms. The analytical solutions give synthetic biologists some clues to design gene oscillators with robust and desired period.
Figures
Similar articles
-
Cyclic negative feedback systems: what is the chance of oscillation?Bull Math Biol. 2014 May;76(5):1155-93. doi: 10.1007/s11538-014-9959-1. Epub 2014 Apr 23. Bull Math Biol. 2014. PMID: 24756857
-
Analytical approximations for the amplitude and period of a relaxation oscillator.BMC Syst Biol. 2009 Jan 14;3:6. doi: 10.1186/1752-0509-3-6. BMC Syst Biol. 2009. PMID: 19144174 Free PMC article.
-
Quasi-multiparameter sensitivity measure for robustness analysis of complex biochemical networks.J Theor Biol. 2011 Mar 7;272(1):174-86. doi: 10.1016/j.jtbi.2010.12.012. Epub 2010 Dec 14. J Theor Biol. 2011. PMID: 21163268
-
Mammalian circadian signaling networks and therapeutic targets.Nat Chem Biol. 2007 Oct;3(10):630-9. doi: 10.1038/nchembio.2007.37. Nat Chem Biol. 2007. PMID: 17876320 Review.
-
Design principles of biochemical oscillators.Nat Rev Mol Cell Biol. 2008 Dec;9(12):981-91. doi: 10.1038/nrm2530. Epub 2008 Oct 30. Nat Rev Mol Cell Biol. 2008. PMID: 18971947 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

