Design of a novel metal binding peptide by molecular dynamics simulation to sequester Cu and Zn ions
- PMID: 25598801
- PMCID: PMC4292183
Design of a novel metal binding peptide by molecular dynamics simulation to sequester Cu and Zn ions
Abstract
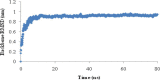
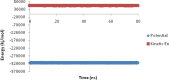
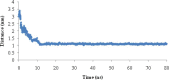
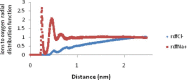
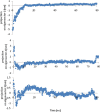
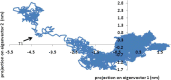
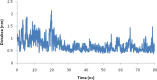
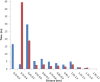
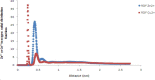
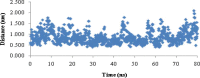
Heavy metal toxicity has serious adverse effects on the environment. The metal sequestering characteristics of a novel metal binding peptide (Glu-Cys)11 Gly+linker+hexahistidine (EC11:His6) was investigated to determine if it can absorb Cu(2+) or Zn(2+) cations. Molecular dynamics simulations were carried out using a model of 6 Cu(2+) or Zn(2+) and other ions enclosed in a fully hydrated simulation box with the designed peptide. Totally, 240 nano second (ns) simulations were done in three phases. Results showed that the selected linker is able to separate two domains of this peptide and that the carboxyl oxygens of Glu residues of EC11 in the designed peptide can absorb these ions. Sequestration of Cu(2+) or Zn(2+) ions by the designed peptide does not change overall tertiary and secondary structures of peptide.
Keywords: Metal binding peptide; Molecular dynamics.
Figures


Similar articles
-
Heterogeneous behavior of metalloproteins toward metal ion binding and selectivity: insights from molecular dynamics studies.J Biomol Struct Dyn. 2016 Jul;34(7):1470-85. doi: 10.1080/07391102.2015.1080629. Epub 2015 Sep 2. J Biomol Struct Dyn. 2016. PMID: 26248730
-
Accelerated Molecular Dynamics to Explore the Binding of Transition Metals to Amyloid-β.ACS Chem Neurosci. 2021 Nov 3;12(21):4065-4075. doi: 10.1021/acschemneuro.1c00466. Epub 2021 Oct 20. ACS Chem Neurosci. 2021. PMID: 34669379
-
Probing the binding of Cu(2+) ions to a fragment of the Aβ(1-42) polypeptide using fluorescence spectroscopy, isothermal titration calorimetry and molecular dynamics simulations.Biophys Chem. 2016 Sep;216:44-50. doi: 10.1016/j.bpc.2016.06.006. Epub 2016 Jul 2. Biophys Chem. 2016. PMID: 27398680
-
Cu and Zn interactions with Aβ peptides: consequence of coordination on aggregation and formation of neurotoxic soluble Aβ oligomers.Metallomics. 2019 Jan 23;11(1):64-84. doi: 10.1039/c8mt00203g. Metallomics. 2019. PMID: 30234208 Review.
-
Peptide folding, metal-binding mechanisms, and binding site structures in metallothioneins.Exp Biol Med (Maywood). 2006 Oct;231(9):1488-99. doi: 10.1177/153537020623100907. Exp Biol Med (Maywood). 2006. PMID: 17018871 Review.
Cited by
-
Localization of Zn2+ ions affects the structural folding and mechanics of Nereis virens Nvjp-1.Soft Matter. 2023 May 31;19(21):3917-3924. doi: 10.1039/d3sm00360d. Soft Matter. 2023. PMID: 37199087 Free PMC article.
-
In silico design of two novel fusion proteins, p28-IL-24 and p28-M4, targeted to breast cancer cells.Res Pharm Sci. 2020 May 11;15(2):200-208. doi: 10.4103/1735-5362.283820. eCollection 2020 Apr. Res Pharm Sci. 2020. PMID: 32582360 Free PMC article.
-
Theoretical design of a new chimeric protein for the treatment of breast cancer.Res Pharm Sci. 2016 May-Jun;11(3):187-99. Res Pharm Sci. 2016. PMID: 27499788 Free PMC article.
-
A theoretical and experimental study of calcium, iron, zinc, cadmium, and sodium ions absorption by aspartame.J Biol Phys. 2017 Mar;43(1):87-103. doi: 10.1007/s10867-016-9435-2. Epub 2017 Feb 1. J Biol Phys. 2017. PMID: 28150114 Free PMC article.
-
Investigation of therapeutic potential of the Il24-p20 fusion protein against breast cancer: an in-silico approach.In Silico Pharmacol. 2024 Sep 18;12(2):84. doi: 10.1007/s40203-024-00252-x. eCollection 2024. In Silico Pharmacol. 2024. PMID: 39301086
References
-
- Broadley MR, White PJ, Hammond JP. Zinc in plants. New Phytol. 2007;173:677–702. - PubMed
-
- Sugarman B. Zinc and Infection. Rev Infect Dis. 1983;5:137–147. - PubMed
-
- Russell R, Beard JL, Cousins RJ. Washington DC: National Academies Press; 2001. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc. - PubMed
-
- Greenwood NN, Earnshaw A, Earnshaw AN. 2nd ed. Oxford: Butterworth-Heinemann; 1997. Chemistry of the Elements; p. 1202.
LinkOut - more resources
Full Text Sources
Miscellaneous
