Initiation of protein-primed picornavirus RNA synthesis
- PMID: 25592245
- PMCID: PMC4476921
- DOI: 10.1016/j.virusres.2014.12.028
Initiation of protein-primed picornavirus RNA synthesis
Abstract
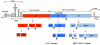
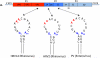
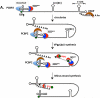
Plus strand RNA viruses use different mechanisms to initiate the synthesis of their RNA chains. The Picornaviridae family constitutes a large group of plus strand RNA viruses that possess a small terminal protein (VPg) covalently linked to the 5'-end of their genomes. The RNA polymerases of these viruses use VPg as primer for both minus and plus strand RNA synthesis. In the first step of the initiation reaction the RNA polymerase links a UMP to the hydroxyl group of a tyrosine in VPg using as template a cis-replicating element (cre) positioned in different regions of the viral genome. In this review we will summarize what is known about the initiation reaction of protein-primed RNA synthesis by the RNA polymerases of the Picornaviridae. As an example we will use the RNA polymerase of poliovirus, the prototype of Picornaviridae. We will also discuss models of how these nucleotidylylated protein primers might be used, together with viral and cellular replication proteins and other cis-replicating RNA elements, during minus and plus strand RNA synthesis.
Keywords: Cis-replicating RNA element (cre); Picornavirus; RNA polymerase; RNA replication; Terminal protein VPg; Uridylylation of VPg.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures



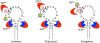

Similar articles
-
Formation and working mechanism of the picornavirus VPg uridylylation complex.Curr Opin Virol. 2014 Dec;9:24-30. doi: 10.1016/j.coviro.2014.09.003. Epub 2014 Sep 19. Curr Opin Virol. 2014. PMID: 25240314 Review.
-
Poliovirus cis-acting replication element-dependent VPg Uridylylation lowers the Km of the initiating nucleoside triphosphate for viral RNA replication.J Virol. 2008 Oct;82(19):9400-8. doi: 10.1128/JVI.00427-08. Epub 2008 Jul 23. J Virol. 2008. PMID: 18653453 Free PMC article.
-
Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis.J Virol. 2003 Apr;77(8):4739-50. doi: 10.1128/jvi.77.8.4739-4750.2003. J Virol. 2003. PMID: 12663781 Free PMC article.
-
Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg.J Virol. 2000 Nov;74(22):10359-70. doi: 10.1128/jvi.74.22.10359-10370.2000. J Virol. 2000. PMID: 11044080 Free PMC article.
-
Cis-active RNA elements (CREs) and picornavirus RNA replication.Virus Res. 2009 Feb;139(2):240-52. doi: 10.1016/j.virusres.2008.07.027. Epub 2008 Sep 20. Virus Res. 2009. PMID: 18773930 Free PMC article. Review.
Cited by
-
Conservation of the HBV RNA element epsilon in nackednaviruses reveals ancient origin of protein-primed reverse transcription.Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2022373118. doi: 10.1073/pnas.2022373118. Proc Natl Acad Sci U S A. 2021. PMID: 33753499 Free PMC article.
-
Tolerance of Senecavirus A to Mutations in Its Kissing-Loop or Pseudoknot Structure Computationally Predicted in 3' Untranslated Region.Front Microbiol. 2022 May 30;13:889480. doi: 10.3389/fmicb.2022.889480. eCollection 2022. Front Microbiol. 2022. PMID: 35707163 Free PMC article.
-
Insights into the Functions of eIF4E-Biding Motif of VPg in Potato Virus A Infection.Viruses. 2020 Feb 11;12(2):197. doi: 10.3390/v12020197. Viruses. 2020. PMID: 32053987 Free PMC article.
-
Picornavirus 2C proteins: structure-function relationships and interactions with host factors.Front Cell Infect Microbiol. 2024 Feb 23;14:1347615. doi: 10.3389/fcimb.2024.1347615. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38465233 Free PMC article. Review.
-
Elusive Trans-Acting Factors Which Operate with Type I (Poliovirus-like) IRES Elements.Int J Mol Sci. 2022 Dec 7;23(24):15497. doi: 10.3390/ijms232415497. Int J Mol Sci. 2022. PMID: 36555135 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

