Essential roles of leucine-rich glioma inactivated 1 in the development of embryonic and postnatal cerebellum
- PMID: 25591666
- PMCID: PMC4296302
- DOI: 10.1038/srep07827
Essential roles of leucine-rich glioma inactivated 1 in the development of embryonic and postnatal cerebellum
Abstract
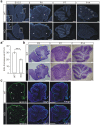
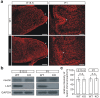
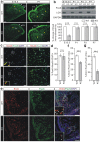
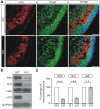
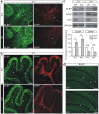
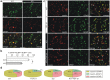
Leucine-rich glioma inactivated 1 (LGI1) is a secreted protein that interacts with ADAM transmembrane proteins, and its mutations are linked to human epilepsy. The function of LGI1 in CNS development remains undefined. Here, we report novel functions of LGI1 in the generation of cerebellar granule precursors (CGPs) and differentiation of radial glial cells (RGCs) in the cerebellum. A reduction in external granule layer thickness and defects in foliation were seen in embryonic and new-born LGI1 knockout (KO) mice. BrdU staining showed an inhibited proliferation of CGPs in KO embryos, which might be explained by the reduced Sonic hedgehog in embryos. In addition, the differentiation of RGCs into Bergmann glias was suppressed in KO mice. Enhanced Jagged1-Notch1 signaling in KO mice via reduced β-secretase proteolysis suggests that altered phenotype of RGCs is due to abnormal Notch1 signaling. Together, our results demonstrate that LGI1 is an essential player in the cerebellar development.
Figures

Similar articles
-
LGI1 is involved in the development of mouse brain.Cerebellum. 2015 Feb;14(1):12-4. doi: 10.1007/s12311-014-0628-6. Cerebellum. 2015. PMID: 25471260 Review.
-
Abnormal cerebellar development and Purkinje cell defects in Lgl1-Pax2 conditional knockout mice.Dev Biol. 2014 Nov 1;395(1):167-81. doi: 10.1016/j.ydbio.2014.07.007. Epub 2014 Jul 19. Dev Biol. 2014. PMID: 25050931
-
Leucine-rich glioma inactivated 1 (Lgi1), an epilepsy-related secreted protein, has a nuclear localization signal and localizes to both the cytoplasm and the nucleus of the caudal ganglionic eminence neurons.Eur J Neurosci. 2012 Aug;36(3):2284-92. doi: 10.1111/j.1460-9568.2012.08129.x. Epub 2012 May 22. Eur J Neurosci. 2012. PMID: 22612501
-
Suppressor of fused controls cerebellar neuronal differentiation in a manner modulated by GLI3 repressor and Fgf15.Dev Dyn. 2018 Jan;247(1):156-169. doi: 10.1002/dvdy.24526. Epub 2017 Jun 15. Dev Dyn. 2018. PMID: 28560839
-
Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling.Development. 2004 Jul;131(13):3159-68. doi: 10.1242/dev.01188. Development. 2004. PMID: 15197161
Cited by
-
Celecoxib Ameliorates Seizure Susceptibility in Autosomal Dominant Lateral Temporal Epilepsy.J Neurosci. 2018 Mar 28;38(13):3346-3357. doi: 10.1523/JNEUROSCI.3245-17.2018. Epub 2018 Feb 28. J Neurosci. 2018. PMID: 29491011 Free PMC article.
-
MEA6 Deficiency Impairs Cerebellar Development and Motor Performance by Tethering Protein Trafficking.Front Cell Neurosci. 2019 Jun 11;13:250. doi: 10.3389/fncel.2019.00250. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31244610 Free PMC article.
-
A novel LGI1 mutation causing autosomal dominant lateral temporal lobe epilepsy confirmed by a precise knock-in mouse model.CNS Neurosci Ther. 2022 Feb;28(2):237-246. doi: 10.1111/cns.13761. Epub 2021 Nov 12. CNS Neurosci Ther. 2022. PMID: 34767694 Free PMC article.
-
Downregulation of Glutamate Transporter EAAT4 by Conditional Knockout of Rheb1 in Cerebellar Purkinje Cells.Cerebellum. 2016 Jun;15(3):314-21. doi: 10.1007/s12311-015-0701-9. Cerebellum. 2016. PMID: 26194056
-
A Potent Antagonist of Smoothened in Hedgehog Signaling for Epilepsy.Int J Mol Sci. 2022 Nov 22;23(23):14505. doi: 10.3390/ijms232314505. Int J Mol Sci. 2022. PMID: 36498832 Free PMC article.
References
-
- Gu W., Brodtkorb E. & Steinlein O. K. LGI1 is mutated in familial temporal lobe epilepsy characterized by aphasic seizures. Ann. Neurol. 52, 364–367 (2002). - PubMed
-
- Senechal K. R., Thaller C. & Noebels J. L. ADPEAF mutations reduce levels of secreted LGI1, a putative tumor suppressor protein linked to epilepsy. Hum. Mol. Genet. 14, 1613–1620 (2005). - PubMed
-
- Fukata Y. et al. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 313, 1792–1795 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

