A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms
- PMID: 25588944
- PMCID: PMC4331008
- DOI: 10.15252/embj.201490062
A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms
Abstract
The loading of small interfering RNAs (siRNAs) and microRNAs into Argonaute proteins is enhanced by Hsp90 and ATP in diverse eukaryotes. However, whether this loading also occurs independently of Hsp90 and ATP remains unclear. We show that the Tetrahymena Hsp90 co-chaperone Coi12p promotes siRNA loading into the Argonaute protein Twi1p in both ATP-dependent and ATP-independent manners in vitro. The ATP-dependent activity requires Hsp90 and the tetratricopeptide repeat (TPR) domain of Coi12p, whereas these factors are dispensable for the ATP-independent activity. Both activities facilitate siRNA loading by counteracting the Twi1p-binding protein Giw1p, which is important to specifically sort the 26- to 32-nt siRNAs to Twi1p. Although Coi12p lacking its TPR domain does not bind to Hsp90, it can partially restore the siRNA loading and DNA elimination defects of COI12 knockout cells, suggesting that Hsp90- and ATP-independent loading of siRNA occurs in vivo and plays a physiological role in Tetrahymena.
Keywords: Argonaute protein; RISC assembly; RNAi; Tetrahymena; siRNA.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

Life cycle of Tetrahymena. A single cell of Tetrahymena thermophila contains two different types of nuclei: a macronucleus (MAC) and a micronucleus (MIC). When sufficient nutrients are available, Tetrahymena grows by binary fission, and the MAC and MIC are divided independently (vegetative growth, green). After prolonged starvation (blue), two cells of complementary mating types fuse to begin the sexual reproduction process (conjugation, pink). Their MICs undergo meiosis (a), and one of the meiotic products survives and divides mitotically, giving rise to two pronuclei: one stationary and one migratory (b). The migratory gametic nucleus crosses the conjugation bridge (c) and fuses with the stationary nucleus to produce the zygotic nucleus (d). The zygotic nucleus divides twice (e). Two of the products differentiate into the new MACs, whereas the other two remain as MICs (f). The parental MAC is degraded, and the pair is dissolved (g). The exconjugants resume vegetative growth when nutrients are available (h). The approximate time at which each event occurs in our culture condition is indicated (hpm: hours post-mixing).
The expression of the predicted Tetrahymena genes showing more than 19 expressed sequence tags (ESTs) from conjugating cells but not vegetative cells was analyzed by RT–PCR using total RNAs from exponentially growing vegetative cells (“E”); 16-h starved cells (“S”); or conjugating cells at 2, 4, 6, 8, 10, and 12 hpm. Twenty-two genes that were exclusively expressed during conjugation are marked with a yellow background. Eleven genes that were analyzed by gene knockout (KO) are indicated in the “KO” column. The phenotypes are described in the “KO phenotype” column. The predicted TTHERM_00526270 contains two genes, which were analyzed separately in this study.
Exconjugants of wild-type (WT) and KO strains for the indicated COI genes at 36–48 hpm were used to detect Tlr1 IES elements by fluorescence in situ hybridization (FISH). The percentages of total exconjugants that showed severe (++), mild (+), and no (−) DNA elimination defects are given (n > 100). Representative pictures of exconjugants showing each phenotype are shown above. The Tlr1 FISH signal is in red, and the DNA was stained with DAPI (blue). The MICs (i) and the new MACs (na) are marked.
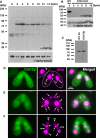
Proteins extracted from wild-type cells at the indicated time point of conjugation (hpm) were analyzed by Western blot with the newly established anti-Coi12p antibody (top) and with an anti-alpha-tubulin antibody (bottom). The positions of the protein molecular weight markers are indicated at the left.
Proteins extracted from wild-type cells at 2 hpm and from the COI12KO cells at the indicated time of conjugation (hpm) were analyzed by Western blot as described in (A).
Proteins extracted from wild-type cells at 4 hpm and recombinantly expressed Coi12p from E. coli were analyzed by Western blot as described in (A).
Immunofluorescence staining of wild-type cells using the anti-Coi12p antibody. Coi12p localizes to the cytoplasm and the MAC in early stage (D, meiotic prophase), mid-stage (E, nuclear exchange), and late stage (F, nuclear alignment) of conjugation. The MICs (i), parental MACs (a) and newly developed MACs (na) are marked with arrowheads. Vegetative (V) and conjugating (C) cells are circled with dotted lines in (D).
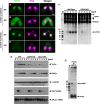
The localization of Twi1p in wild-type (A, C) and COI12KO (B, D) cells at the early (meiotic prophase; A, B) and mid- (nuclear exchange; C, D) conjugation stages was analyzed by immunofluorescence staining using an anti-Twi1p antibody. Twi1p was localized in the parental MACs at both stages in wild-type cells (A, C). In contrast, in COI12KO cells, Twi1p was localized in the cytoplasm at the early stage (B) and became undetectable at the mid-stage (D). All pictures share the scale bar (10 μm) shown in (A).
The expression of Twi1p in wild-type, COI12KO, and DCL1KO cells at the indicated time points of conjugation was analyzed by Western blot using an anti-Twi1p antibody (top panel). The expression of Pdd1p was analyzed using an anti-Pdd1p antibody as an indicator of the proper progress of conjugation (2nd panel), and the expression of alpha-tubulin was analyzed by anti-alpha-tubulin antibody as a loading control (3rd panel). The expression of TWI1mRNA (4th panel) and RPL21mRNA (bottom panel) was analyzed by Northern blot.
Total RNA was extracted from wild-type and COI12KO cells at the indicated time points of conjugation, separated on a denaturing gel, and visualized by nucleic acid staining (GelRed). Bands corresponding to rRNAs, tRNAs, and scnRNAs are marked with an arrowhead.
Twi1p was immunoprecipitated with an anti-Twi1p antibody from wild-type (WT) and COI12KO (KO) cells at 2 hpm. Co-precipitated RNAs were separated on a denaturing gel and visualized by the nucleic acid dye GelRed. The position of scnRNAs is marked with an arrowhead. The sizes of RNA markers (M) are indicated at the left. Precipitated Twi1p was analyzed by Western blot using an anti-Twi1p antibody (bottom).
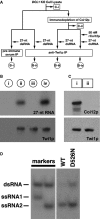
A schematic drawing of the scnRNA loading assays in cell lysates. The circled samples (B-i, B-ii, B-iii, and B-iv) correspond to the lanes shown in (B). The boxed samples (C-i, C-ii) correspond to the lanes shown in (C). See text for details.
The 27-nt RNAs that co-precipitated with Twi1p from lysates with the different conditions shown in (A) were separated on a denaturing gel and detected by autoradiography (top). Precipitated Twi1p was detected by Western blot using an anti-Twi1p antibody (bottom).
Proteins in the cell lysate before (i) and after (ii) immunodepletion of Coi12p were analyzed by Western blot using anti-Coi12p (top) and anti-Twi1p (bottom) antibodies.
Cell lysate from the cells expressing FLAG-HA-tagged wild-type Twi1p (WT) or Twi1p in which the aspartic acid 526 was replaced with asparagine (D526N) at 3 hpm was incubated with radiolabeled double-stranded 27-nt RNAs. The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, co-precipitated RNAs were separated on a native gel, and 27-nt RNAs were detected by autoradiography. As markers, ssRNA1 and ssRNA2, which were used for producing the radiolabeled double-stranded 27-nt RNAs, were mixed in 2:1 (left) or 1:2 (right) ratio, allowed to form double-stranded RNAs, and were separated in the same native gel.

Cell lysate from COI12KO cells at 3 hpm was incubated with radiolabeled double-stranded 27-nt RNAs and the different truncated mutants of Coi12p (schematically shown on the top). The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, co-precipitated RNAs were separated on a denaturing gel, and 27-nt RNAs were detected by autoradiography. The intensities of the bands in the different conditions relative to the wild-type (WT) sample (2nd column) are shown on the top. A representative experiment is shown on the bottom.
Cell lysate from COI12KO cells at 3 hpm was incubated with radiolabeled double-stranded 27-nt RNAs, wild-type (W), or ΔTPR mutant (Δ) of recombinant Coi12p, and with (+) or without (−) ATP and the ATP regeneration system. The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, and co-precipitated 27-nt RNAs were detected by autoradiography. The intensities of the bands in the different conditions relative to the sample with wild-type Coi12p and without an ATP supply (1st column) are shown on the top. A representative experiment is shown on the bottom.
Cell lysate from COI12KO cells at 3 hpm was incubated with (+) or without (−) hexokinase followed by radiolabeled double-stranded 27-nt RNAs and recombinant wild-type Coi12p. Loaded RNA was analyzed as in (B). The intensities of the bands relative to the sample without hexokinase are shown.
Cell lysate from DCL1KO cells at 3 hpm was incubated with radiolabeled double-stranded 27-nt RNAs and then with (+) or without (−) ATP and the ATP regeneration system, the HSP70 inhibitor PES, and the HSP90 inhibitor 17-AAG. Loaded RNA was analyzed as in (B). The intensities of the bands relative to the sample without ATP and any inhibitors (1st column) are shown on the top.
Cell lysate from COI12KO cells at 3 hpm was incubated with radiolabeled double-stranded 27-nt RNAs, recombinant wild-type Coi12p, with (+) or without (−) ATP, and with (+) or without (−) ATP-γ-S. Loaded RNA was analyzed as in (B). The intensities of the bands relative to the sample with ATP (the 1st lane) are shown on the top.
GST-tagged wild-type Coi12p (GST-Coi12p) or GST-tagged Coi12p lacking the TPR domain (GST-Coi12p-ΔTPR) was incubated with His-tagged wild-type Hsp82p (His-Hsp82p) or His-tagged Hsp82p lacking the C-terminal MEDVD sequence (His-Hsp82p-ΔC) and affinity-purified with glutathione-coupled beads. Proteins before (input) or after (GST pull-down) the purification were analyzed by Western blot using anti-GST (top) and anti-His (bottom) antibodies. The positions of GST-Coi12p, GST-Coi12p-ΔTPR, His-Hsp82p, and His-Coi12p-ΔC are marked with arrowheads.
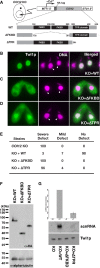
A schematic drawing of the in vivo functional analysis of COI12 mutants. The expression constructs from which HA-tagged Coi12p proteins (wild-type [WT] or the mutant proteins lacking the two FKBDs [ΔFKBD] or the TPR domain [ΔTPR]) are expressed under the control of the cadmium-inducible MTT1 promoter were introduced into the MACs of COI12KO cells.
Cells containing the expression constructs shown in (A) were mated with COI12KO cells in the presence of cadmium ions. The localization of Twi1p was analyzed by immunofluorescence staining using an anti-Twi1p antibody at 3 hpm (B–D). In (B–D), the parental MACs are marked with arrowheads. DNA elimination was analyzed by DNA FISH with probes against Tlr1 IESs at 36 hpm (E). Defects in DNA elimination were categorized as described for Fig1C. The expression of the different Coi12p mutants was analyzed by Western blot using an anti-HA antibody (F, top). Alpha-tubulin was analyzed as a loading control (F, bottom). The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody. Precipitated scnRNA was separated on a denaturing gel and visualized by the nucleic acid staining dye GelRed (G, top and middle). The intensities of bands relative to the sample with the mating expressing wild-type Coi12p (KO+WT) are shown on the top, and a result of a representative experiment is shown in the middle. Precipitated Twi1p was analyzed by Western blot using an anti-Twi1p antibody (bottom). In (G), the standard deviation (SD) between technical replicates is indicated.

The Twi1p-containing complex was immunoprecipitated using an anti-Twi1p antibody from wild-type (W) and DCL1KO (Δ) cells, and input (left) and precipitated (right) proteins were analyzed by Western blot using an anti-Twi1p antibody (top) and an anti-Giw1p antibody (bottom).
Cell lysates from wild-type (WT) or GIW1KO cells at 3 hpm were incubated with radiolabeled double-stranded 27-nt RNAs and with (+) or without (−) ATP and the ATP regeneration system. The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, co-precipitated RNAs were separated on a denaturing gel and detected by autoradiography. The intensities of the bands in the different conditions relative to the sample with wild-type cell lysate and without ATP (1st column) are shown on the top. The standard deviation (SD) between technical replicates is indicated. A result of a representative experiment is shown on the bottom.
RNAs were prepared from cell lysates from wild-type (W) or GIW1KO (Δ) cells with ATP and the ATP regeneration system as in (A) but were separated in a native gel. The positions of double-stranded 27-nt RNAs (dsRNA) and each strand of RNA used to form the 27-nt RNA duplex (ssRNA1 and ssRNA2) are indicated at the left.
Coi12p was immunodepleted from GIW1KO cell lysate at 3 hpm, and the lysate was incubated with radiolabeled double-stranded 27-nt RNAs. As a control, GIW1KO cell lysate without Coi12p depletion was used for the loading assay. The protein samples (i, ii) correspond to the lanes shown in the top right panel. Coi12p and Twi1p in the lysates were detected by Western blot. The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, and co-precipitated 27-nt RNAs were detected by autoradiography. The RNA samples (iii, iv) correspond to the lanes shown in the bottom right panel.
The indicated concentration of MBP-tagged Twi1p (MBP-Twi1p), which was recombinantly expressed in E. coli, was incubated with the radiolabeled double-stranded 27-nt RNAs. Loaded RNA was analyzed by immunoprecipitating MBP-Twi1p with an anti-Twi1p antibody and separated on a denaturing gel, followed by autoradiographic detection.
MBP-Twi1p (50 nM) was incubated with the indicated concentrations of the recombinantly expressed GST-Giw1p and with radiolabeled 27-nt RNA duplexes. Loaded RNA was analyzed as in (C). The intensities of the bands in the different conditions relative to the sample without GST-Giw1p (1st column) are shown on the top. The standard deviation (SD) between technical replicates is indicated. A result of a representative experiment is shown on the bottom.
The indicated concentration of recombinantly expressed GST-tagged Twi1p in which aspartic acid 526 was replaced by asparagine (GST-Twi1p-D526N) or GST alone was incubated with radiolabeled double-stranded 27-nt RNAs. Loaded RNA was analyzed by immunoprecipitating GST-Twi1p-D526N with an anti-Twi1p antibody and separating on a native gel, followed by autoradiographic detection. The positions of double-stranded 27-nt RNAs (dsRNA) and each strand of RNA used to form the 27-nt RNA duplex (ssRNA1 and ssRNA2) are indicated at the right.
Cell lysates from wild-type (WT) or GIW1KO cells at 3 hpm were incubated with a mixture of radiolabeled double-stranded 23-, 27-, 31-, and 35-nt RNAs, ATP, and the ATP regeneration system. The Twi1p-containing complex was immunoprecipitated with an anti-Twi1p antibody, and co-precipitated RNAs were detected by autoradiography. The intensities of each RNA band relative to the 27-nt RNA were normalized to the input sample and are shown on the right. The standard deviation (SD) between technical replicates is indicated. A representative experiment is shown on the left.
Similar articles
-
The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus.Cell. 2010 Mar 5;140(5):692-703. doi: 10.1016/j.cell.2010.02.010. Cell. 2010. PMID: 20211138 Free PMC article.
-
Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins?Wiley Interdiscip Rev RNA. 2016 Sep;7(5):637-60. doi: 10.1002/wrna.1356. Epub 2016 May 16. Wiley Interdiscip Rev RNA. 2016. PMID: 27184117 Free PMC article. Review.
-
Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes.Mol Cell. 2010 Jul 30;39(2):292-9. doi: 10.1016/j.molcel.2010.05.015. Epub 2010 Jun 3. Mol Cell. 2010. PMID: 20605501
-
RNAi-dependent heterochromatin assembly in fission yeast Schizosaccharomyces pombe requires heat-shock molecular chaperones Hsp90 and Mas5.Epigenetics Chromatin. 2018 Jun 4;11(1):26. doi: 10.1186/s13072-018-0199-8. Epigenetics Chromatin. 2018. PMID: 29866182 Free PMC article.
-
The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones.Biochim Biophys Acta. 2012 Mar;1823(3):624-35. doi: 10.1016/j.bbamcr.2011.09.003. Epub 2011 Sep 16. Biochim Biophys Acta. 2012. PMID: 21951723 Review.
Cited by
-
Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes.Nat Commun. 2017 Jun 6;8:15690. doi: 10.1038/ncomms15690. Nat Commun. 2017. PMID: 28585547 Free PMC article.
-
Targeted Gene Disruption by Ectopic Induction of DNA Elimination in Tetrahymena.Genetics. 2015 Sep;201(1):55-64. doi: 10.1534/genetics.115.178525. Epub 2015 Jul 23. Genetics. 2015. PMID: 26205990 Free PMC article.
-
Genes and proteins expressed at different life cycle stages in the model protist Euplotes vannus revealed by both transcriptomic and proteomic approaches.Sci China Life Sci. 2025 Jan;68(1):232-248. doi: 10.1007/s11427-023-2605-9. Epub 2024 Sep 10. Sci China Life Sci. 2025. PMID: 39276255
-
A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena.Mol Biol Cell. 2017 Mar 15;28(6):825-833. doi: 10.1091/mbc.E16-09-0678. Epub 2017 Jan 18. Mol Biol Cell. 2017. PMID: 28100637 Free PMC article.
-
How does Hsp90 function in RNAi-dependent heterochromatin assembly?Curr Genet. 2019 Feb;65(1):87-91. doi: 10.1007/s00294-018-0866-0. Epub 2018 Jul 4. Curr Genet. 2019. PMID: 29974204 Review.
References
-
- Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theor Biol. 1990;143:163–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

