Transcriptional regulation of Sox2 by the retinoblastoma family of pocket proteins
- PMID: 25576924
- PMCID: PMC4413632
- DOI: 10.18632/oncotarget.2996
Transcriptional regulation of Sox2 by the retinoblastoma family of pocket proteins
Abstract
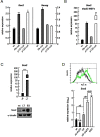
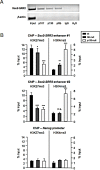
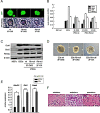
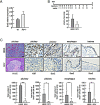
Cellular reprogramming to iPSCs has uncovered unsuspected links between tumor suppressors and pluripotency factors. Using this system, it was possible to identify tumor suppressor p27 as a repressor of Sox2 during differentiation. This led to the demonstration that defects in the repression of Sox2 can contribute to tumor development. The members of the retinoblastoma family of pocket proteins, pRb, p107 and p130, are negative regulators of the cell cycle with tumor suppressor activity and with roles in differentiation. In this work we studied the relative contribution of the retinoblastoma family members to the regulation of Sox2 expression. We found that deletion of Rb or p130 leads to impaired repression of Sox2, a deffect amplified by inactivation of p53. We also identified binding of pRb and p130 to an enhancer with crucial regulatory activity on Sox2 expression. Using cellular reprogramming we tested the impact of the defective repression of Sox2 and confirmed that Rb deficiency allows the generation of iPSCs in the absence of exogenous Sox2. Finally, partial depletion of Sox2 positive cells reduced the pituitary tumor development initiated by Rb loss in vivo. In summary, our results show that Sox2 repression by pRb is a relevant mechanism of tumor suppression.
Figures
Similar articles
-
RB, p130 and p107 differentially repress G1/S and G2/M genes after p53 activation.Nucleic Acids Res. 2019 Dec 2;47(21):11197-11208. doi: 10.1093/nar/gkz961. Nucleic Acids Res. 2019. PMID: 31667499 Free PMC article.
-
Retinoblastoma/p107/p130 pocket proteins: protein dynamics and interactions with target gene promoters.J Biol Chem. 2009 Jul 17;284(29):19265-71. doi: 10.1074/jbc.M808740200. Epub 2009 Mar 11. J Biol Chem. 2009. PMID: 19279001 Free PMC article.
-
Retinoblastoma protein (pRb), but not p107 or p130, is required for maintenance of enterocyte quiescence and differentiation in small intestine.J Biol Chem. 2009 Jan 2;284(1):134-140. doi: 10.1074/jbc.M806133200. Epub 2008 Nov 3. J Biol Chem. 2009. PMID: 18981186 Free PMC article.
-
Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review.
-
From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway.J Cell Biochem. 2007 Dec 15;102(6):1400-4. doi: 10.1002/jcb.21609. J Cell Biochem. 2007. PMID: 17979151 Review.
Cited by
-
Peroxisome proliferator-activated receptor-β/δ inhibits human neuroblastoma cell tumorigenesis by inducing p53- and SOX2-mediated cell differentiation.Mol Carcinog. 2017 May;56(5):1472-1483. doi: 10.1002/mc.22607. Epub 2017 Jan 13. Mol Carcinog. 2017. PMID: 27996177 Free PMC article.
-
CMV70-3P miRNA contributes to the CMV mediated glioma stemness and represents a target for glioma experimental therapy.Oncotarget. 2017 Apr 18;8(16):25989-25999. doi: 10.18632/oncotarget.11175. Oncotarget. 2017. PMID: 27517625 Free PMC article.
-
Context-Dependent Impact of RAS Oncogene Expression on Cellular Reprogramming to Pluripotency.Stem Cell Reports. 2019 May 14;12(5):1099-1112. doi: 10.1016/j.stemcr.2019.04.006. Epub 2019 May 2. Stem Cell Reports. 2019. PMID: 31056476 Free PMC article.
-
SOX2 is required independently in both stem and differentiated cells for pituitary tumorigenesis in p27-null mice.Proc Natl Acad Sci U S A. 2021 Feb 16;118(7):e2017115118. doi: 10.1073/pnas.2017115118. Proc Natl Acad Sci U S A. 2021. PMID: 33574062 Free PMC article.
-
Retinoblastoma Protein Paralogs and Tumor Suppression.Front Genet. 2022 Mar 18;13:818719. doi: 10.3389/fgene.2022.818719. eCollection 2022. Front Genet. 2022. PMID: 35368709 Free PMC article. Review.
References
-
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harbor symposia on quantitative biology. 2008;73:171–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

