RNA-sequencing analysis of Trichophyton rubrum transcriptome in response to sublethal doses of acriflavine
- PMID: 25573029
- PMCID: PMC4243288
- DOI: 10.1186/1471-2164-15-S7-S1
RNA-sequencing analysis of Trichophyton rubrum transcriptome in response to sublethal doses of acriflavine
Abstract
Background: The dermatophyte Trichophyton rubrum is an anthropophilic filamentous fungus that infects keratinized tissues and is the most common etiologic agent isolated in human dermatophytoses. The clinical treatment of these infections is challenging because only few antifungal drugs are commercially available. To understand the mode of action of cytotoxic drugs against fungi, we evaluated the time-dependent effects of acriflavine on T. rubrum transcriptome using high-throughput RNA-sequencing (RNA-seq) technology.
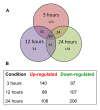
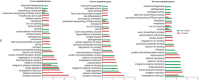
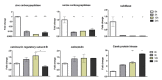
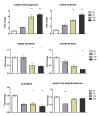
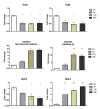
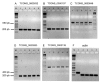
Results: RNA-seq analysis generated approximately 200 million short reads that were mapped to the Broad Institute's Dermatophyte Comparative Database before differential gene expression analysis was performed. By employing a stringent cut-off threshold of -1.5 and 1.5 log₂-fold changes in gene expression, a subset of 490 unique genes were found to be modulated in T. rubrum in response to acriflavine exposure. Among the selected genes, 69 genes were modulated at all exposure time points. Functional categorization indicated the putative involvement of these genes in various cellular processes such as oxidation-reduction reaction, transmembrane transport, and metal ion binding. Interestingly, genes putatively involved in the pathogenicity of dermatophytoses were down-regulated suggesting that this drug interferes with the virulence of T. rubrum. Moreover, we identified 159 novel putative transcripts in intergenic regions and two transcripts in intron regions of T. rubrum genome.
Conclusion: The results provide insights into the molecular events underlying the stress responses of T. rubrum to acriflavine, revealing that this drug interfered with important molecular events involved in the establishment and maintenance of fungal infection in the host. In addition, the identification of novel transcripts will further enable the improvement of gene annotation and open reading frame prediction of T. rubrum and other dermatophyte genomes.
Figures
Similar articles
-
Over-expression of genes coding for proline oxidase, riboflavin kinase, cytochrome c oxidase and an MFS transporter induced by acriflavin in Trichophyton rubrum.Med Mycol. 2008 Mar;46(2):135-9. doi: 10.1080/13693780701742381. Med Mycol. 2008. PMID: 18324492
-
Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum.BMC Microbiol. 2010 Feb 8;10:39. doi: 10.1186/1471-2180-10-39. BMC Microbiol. 2010. PMID: 20144196 Free PMC article.
-
Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern.BMC Genomics. 2016 Mar 18;17:249. doi: 10.1186/s12864-016-2567-8. BMC Genomics. 2016. PMID: 26993619 Free PMC article.
-
Back to the future for dermatophyte genomics.mBio. 2012 Oct 30;3(6):e00381-12. doi: 10.1128/mBio.00381-12. mBio. 2012. PMID: 23111872 Free PMC article. Review.
-
State-of-the-Art Dermatophyte Infections: Epidemiology Aspects, Pathophysiology, and Resistance Mechanisms.J Fungi (Basel). 2021 Aug 3;7(8):629. doi: 10.3390/jof7080629. J Fungi (Basel). 2021. PMID: 34436168 Free PMC article. Review.
Cited by
-
Evaluation of the antidermatophytic activity of potassium salts of N-acylhydrazinecarbodithioates and their aminotriazole-thione derivatives.Sci Rep. 2024 Feb 12;14(1):3521. doi: 10.1038/s41598-024-54025-9. Sci Rep. 2024. PMID: 38347115 Free PMC article.
-
Molecular characterization of carbendazim resistance of Fusarium species complex that causes sugarcane pokkah boeng disease.BMC Genomics. 2019 Feb 7;20(1):115. doi: 10.1186/s12864-019-5479-6. BMC Genomics. 2019. PMID: 30732567 Free PMC article.
-
Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms.BMC Genomics. 2015 Oct 24;16:855. doi: 10.1186/s12864-015-2043-x. BMC Genomics. 2015. PMID: 26499483 Free PMC article.
-
Acriflavine, an Acridine Derivative for Biomedical Application: Current State of the Art.J Med Chem. 2022 Sep 8;65(17):11415-11432. doi: 10.1021/acs.jmedchem.2c00573. Epub 2022 Aug 26. J Med Chem. 2022. PMID: 36018000 Free PMC article. Review.
-
Reassessing the Use of Undecanoic Acid as a Therapeutic Strategy for Treating Fungal Infections.Mycopathologia. 2021 Jun;186(3):327-340. doi: 10.1007/s11046-021-00550-4. Epub 2021 Apr 9. Mycopathologia. 2021. PMID: 33835367 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

