The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts
- PMID: 25569378
- PMCID: PMC4615894
- DOI: 10.1080/19491034.2014.1003509
The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts
Abstract
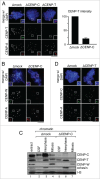
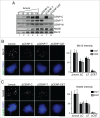
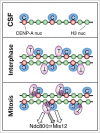
The centromere is the chromosomal region in which the kinetochore is assembled to orchestrate chromosome segregation. It is defined by the presence of a histone H3 variant called Centromere Protein A (CENP-A) or CenH3. Propagation of centromere identity entails deposition of new CENP-A upon exit from mitosis in vertebrate cells. A group of 16 proteins that co-immunoprecipitate with CENP-A, the Constitutive Centromere Associated Network or CCAN, contribute to kinetochore assembly and function. For most of them it is still unclear how and when they are recruited to centromeres and whether they have a role in CENP-A deposition. Taking advantage of the Xenopus egg cell-free system, we have addressed these issues for CCAN proteins CENP-C, CENP-T and CENP-W. CENP-C recruitment occurs as soon as sperm DNA, containing CENP-A, is added to the egg extract, and continues after de novo incorporation of CENP-A in early interphase. In contrast, centromeric recruitment of CENP-T occurs in late interphase and precedes that of CENP-W, which occurs in mitosis. Unlike CENP-C, CENP-T and CENP-W do not participate in CENP-A deposition. However, like CENP-C, they play a major role in kinetochore assembly. Depletion of CENP-C results in reduced amount of CENP-T at centromeres, an effect more prominent in mitosis than in interphase. In spite of this, kinetochores can still be assembled under this condition although the recruitment of Ndc80 and Mis12 is decreased. Our results support the existence of 2 pathways for kinetochore assembly directed by CENP-C and CENP-T/W, which can be reconstituted in Xenopus egg extracts.
Keywords: CCAN; CCAN, Constitutive Centromere Associated Network; CENP, centromere protein; CENP-A; CenH3; kinetochore.
Figures
Similar articles
-
Dissection of CENP-C-directed centromere and kinetochore assembly.Mol Biol Cell. 2009 Oct;20(19):4246-55. doi: 10.1091/mbc.e09-05-0378. Epub 2009 Jul 29. Mol Biol Cell. 2009. PMID: 19641019 Free PMC article.
-
In vitro centromere and kinetochore assembly on defined chromatin templates.Nature. 2011 Aug 28;477(7364):354-8. doi: 10.1038/nature10379. Nature. 2011. PMID: 21874020 Free PMC article.
-
CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore.Cell. 2008 Dec 12;135(6):1039-52. doi: 10.1016/j.cell.2008.10.019. Cell. 2008. PMID: 19070575
-
The ABCs of CENPs.Chromosoma. 2011 Oct;120(5):425-46. doi: 10.1007/s00412-011-0330-0. Epub 2011 Jul 13. Chromosoma. 2011. PMID: 21751032 Review.
-
Conserved and divergent mechanisms of inner kinetochore assembly onto centromeric chromatin.Curr Opin Struct Biol. 2023 Aug;81:102638. doi: 10.1016/j.sbi.2023.102638. Epub 2023 Jun 20. Curr Opin Struct Biol. 2023. PMID: 37343495 Review.
Cited by
-
Permitted and restricted steps of human kinetochore assembly in mitotic cell extracts.Mol Biol Cell. 2021 Jun 15;32(13):1241-1255. doi: 10.1091/mbc.E20-07-0461. Epub 2021 May 6. Mol Biol Cell. 2021. PMID: 33956511 Free PMC article.
-
Kinetochore assembly and function through the cell cycle.Chromosoma. 2016 Sep;125(4):645-59. doi: 10.1007/s00412-016-0608-3. Epub 2016 Jul 4. Chromosoma. 2016. PMID: 27376724 Review.
-
Bub1 targeting to centromeres is sufficient for Sgo1 recruitment in the absence of kinetochores.Chromosoma. 2017 Mar;126(2):279-286. doi: 10.1007/s00412-016-0592-7. Epub 2016 Apr 26. Chromosoma. 2017. PMID: 27116032 Free PMC article.
-
Distinct Roles of the Chromosomal Passenger Complex in the Detection of and Response to Errors in Kinetochore-Microtubule Attachment.Dev Cell. 2017 Sep 25;42(6):640-654.e5. doi: 10.1016/j.devcel.2017.08.022. Dev Cell. 2017. PMID: 28950102 Free PMC article.
-
Insights from the reconstitution of the divergent outer kinetochore of Drosophila melanogaster.Open Biol. 2016 Feb;6(2):150236. doi: 10.1098/rsob.150236. Open Biol. 2016. PMID: 26911624 Free PMC article.
References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008; 9:33-46; PMID:18097444; http://dx.doi.org/10.1038/nrm2310 - DOI - PubMed
-
- Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J 2009; 28:2511-31; PMID:19629042; http://dx.doi.org/10.1038/emboj.2009.173 - DOI - PMC - PubMed
-
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 2003; 112:407-21; PMID:12600307; http://dx.doi.org/10.1016/S0092-8674(03)00115-6 - DOI - PubMed
-
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 2008; 9:923-37; PMID:19002142; http://dx.doi.org/10.1038/nrg2466 - DOI - PMC - PubMed
-
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2002; 2:319-30; PMID:11879637; http://dx.doi.org/10.1016/S1534-5807(02)00135-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
