Polycomb genes, miRNA, and their deregulation in B-cell malignancies
- PMID: 25568352
- PMCID: PMC4335077
- DOI: 10.1182/blood-2014-10-606822
Polycomb genes, miRNA, and their deregulation in B-cell malignancies
Abstract
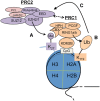
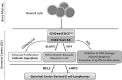
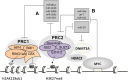
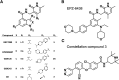
Posttranslational modifications of histone proteins represent a fundamental means to define distinctive epigenetic states and regulate gene expression during development and differentiation. Aberrations in various chromatin-modulation pathways are commonly used by tumors to initiate and maintain oncogenesis, including lymphomagenesis. Recently, increasing evidence has demonstrated that polycomb group (PcG) proteins, a subset of histone-modifying enzymes known to be crucial for B-cell maturation and differentiation, play a central role in malignant transformation of B cells. PcG hyperactivity in B-cell lymphomas is caused by overexpression or recurrent mutations of PcG genes and deregulation of microRNAs (miRNAs) or transcription factors such as c-MYC, which regulate PcG expression. Interplays of PcG and miRNA deregulations often establish a vicious signal-amplification loop in lymphoma associated with adverse clinical outcomes. Importantly, aberrant enzymatic activities associated with polycomb deregulation, notably those caused by EZH2 gain-of-function mutations, have provided a rationale for developing small-molecule inhibitors as novel therapies. In this review, we summarize our current understanding of PcG-mediated gene silencing, interplays of PcG with other epigenetic regulators such as miRNAs during B-cell differentiation and lymphomagenesis, and recent advancements in targeted strategies against PcG as promising therapeutics for B-cell malignancies.
© 2015 by The American Society of Hematology.
Figures
Similar articles
-
Epigenetic regulators: Polycomb-miRNA circuits in cancer.Biochim Biophys Acta. 2016 May;1859(5):697-704. doi: 10.1016/j.bbagrm.2016.03.005. Epub 2016 Mar 11. Biochim Biophys Acta. 2016. PMID: 26975854 Review.
-
The roles of Polycomb group proteins in hematopoietic stem cells and hematological malignancies.Int J Hematol. 2016 Jun;103(6):634-42. doi: 10.1007/s12185-016-2011-5. Epub 2016 Apr 16. Int J Hematol. 2016. PMID: 27086351 Review.
-
Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer.Cell Cycle. 2006 Aug;5(16):1886-901. doi: 10.4161/cc.5.16.3222. Epub 2006 Aug 15. Cell Cycle. 2006. PMID: 16963837
-
Nucleotide substitutions revealing specific functions of Polycomb group genes.Mol Genet Metab. 2015 Apr;114(4):547-56. doi: 10.1016/j.ymgme.2015.01.007. Epub 2015 Jan 30. Mol Genet Metab. 2015. PMID: 25669595 Review.
-
A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis.Blood. 2014 Jun 19;123(25):3914-24. doi: 10.1182/blood-2012-12-473439. Epub 2014 May 6. Blood. 2014. PMID: 24802772
Cited by
-
sRNA-Effector: A tool to expedite discovery of small RNA regulators.iScience. 2024 Feb 20;27(3):109300. doi: 10.1016/j.isci.2024.109300. eCollection 2024 Mar 15. iScience. 2024. PMID: 38469560 Free PMC article.
-
miR-141 is involved in BRD7-mediated cell proliferation and tumor formation through suppression of the PTEN/AKT pathway in nasopharyngeal carcinoma.Cell Death Dis. 2016 Mar 24;7(3):e2156. doi: 10.1038/cddis.2016.64. Cell Death Dis. 2016. PMID: 27010857 Free PMC article.
-
Epigenetic Heterogeneity of B-Cell Lymphoma: DNA Methylation, Gene Expression and Chromatin States.Genes (Basel). 2015 Sep 7;6(3):812-40. doi: 10.3390/genes6030812. Genes (Basel). 2015. PMID: 26371046 Free PMC article.
-
EZH2 overexpression dampens tumor-suppressive signals via an EGR1 silencer to drive breast tumorigenesis.Oncogene. 2020 Nov;39(48):7127-7141. doi: 10.1038/s41388-020-01484-9. Epub 2020 Oct 2. Oncogene. 2020. PMID: 33009487
-
Epigenetic Heterogeneity of B-Cell Lymphoma: Chromatin Modifiers.Genes (Basel). 2015 Oct 21;6(4):1076-112. doi: 10.3390/genes6041076. Genes (Basel). 2015. PMID: 26506391 Free PMC article.
References
-
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. - PubMed
-
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13(9):363–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

