Assembly of viral genomes from metagenomes
- PMID: 25566226
- PMCID: PMC4270193
- DOI: 10.3389/fmicb.2014.00714
Assembly of viral genomes from metagenomes
Abstract
Viral infections remain a serious global health issue. Metagenomic approaches are increasingly used in the detection of novel viral pathogens but also to generate complete genomes of uncultivated viruses. In silico identification of complete viral genomes from sequence data would allow rapid phylogenetic characterization of these new viruses. Often, however, complete viral genomes are not recovered, but rather several distinct contigs derived from a single entity are, some of which have no sequence homology to any known proteins. De novo assembly of single viruses from a metagenome is challenging, not only because of the lack of a reference genome, but also because of intrapopulation variation and uneven or insufficient coverage. Here we explored different assembly algorithms, remote homology searches, genome-specific sequence motifs, k-mer frequency ranking, and coverage profile binning to detect and obtain viral target genomes from metagenomes. All methods were tested on 454-generated sequencing datasets containing three recently described RNA viruses with a relatively large genome which were divergent to previously known viruses from the viral families Rhabdoviridae and Coronaviridae. Depending on specific characteristics of the target virus and the metagenomic community, different assembly and in silico gap closure strategies were successful in obtaining near complete viral genomes.
Keywords: assembly; metagenome; pathogen; viral metagenomics; virome; virus; virus discovery.
Figures

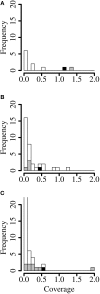
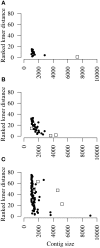
Similar articles
-
FastViromeExplorer-Novel: Recovering Draft Genomes of Novel Viruses and Phages in Metagenomic Data.J Comput Biol. 2023 Apr;30(4):391-408. doi: 10.1089/cmb.2022.0397. Epub 2023 Jan 6. J Comput Biol. 2023. PMID: 36607772
-
VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data.Microbiome. 2017 Jul 6;5(1):69. doi: 10.1186/s40168-017-0283-5. Microbiome. 2017. PMID: 28683828 Free PMC article.
-
Recovering full-length viral genomes from metagenomes.Front Microbiol. 2015 Oct 1;6:1069. doi: 10.3389/fmicb.2015.01069. eCollection 2015. Front Microbiol. 2015. PMID: 26483782 Free PMC article. Review.
-
Evaluating de Novo Assembly and Binning Strategies for Time Series Drinking Water Metagenomes.Microbiol Spectr. 2021 Dec 22;9(3):e0143421. doi: 10.1128/Spectrum.01434-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730411 Free PMC article.
-
Recovering complete and draft population genomes from metagenome datasets.Microbiome. 2016 Mar 8;4:8. doi: 10.1186/s40168-016-0154-5. Microbiome. 2016. PMID: 26951112 Free PMC article. Review.
Cited by
-
ViralCC retrieves complete viral genomes and virus-host pairs from metagenomic Hi-C data.Nat Commun. 2023 Jan 31;14(1):502. doi: 10.1038/s41467-023-35945-y. Nat Commun. 2023. PMID: 36720887 Free PMC article.
-
Microbial and Viral Communities and Their Antibiotic Resistance Genes Throughout a Hospital Wastewater Treatment System.Front Microbiol. 2020 Feb 19;11:153. doi: 10.3389/fmicb.2020.00153. eCollection 2020. Front Microbiol. 2020. PMID: 32140141 Free PMC article.
-
Fragmentation and Coverage Variation in Viral Metagenome Assemblies, and Their Effect in Diversity Calculations.Front Bioeng Biotechnol. 2015 Sep 17;3:141. doi: 10.3389/fbioe.2015.00141. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26442255 Free PMC article.
-
Coding-Complete Genome Sequence of a Pollen-Associated Virus Belonging to the Secoviridae Family Recovered from a Japanese Apricot (Prunus mume) Metagenome Data Set.Microbiol Resour Announc. 2019 Oct 3;8(40):e00881-19. doi: 10.1128/MRA.00881-19. Microbiol Resour Announc. 2019. PMID: 31582454 Free PMC article.
-
Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses.Microbiome. 2018 Jun 28;6(1):119. doi: 10.1186/s40168-018-0507-3. Microbiome. 2018. PMID: 29954453 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

