Modulation of folding energy landscape by charge-charge interactions: linking experiments with computational modeling
- PMID: 25564663
- PMCID: PMC4311858
- DOI: 10.1073/pnas.1410424112
Modulation of folding energy landscape by charge-charge interactions: linking experiments with computational modeling
Abstract
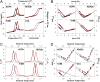
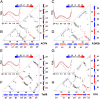
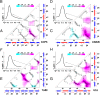
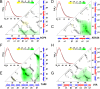
The kinetics of folding-unfolding of a structurally diverse set of four proteins optimized for thermodynamic stability by rational redesign of surface charge-charge interactions is characterized experimentally. The folding rates are faster for designed variants compared with their wild-type proteins, whereas the unfolding rates are largely unaffected. A simple structure-based computational model, which incorporates the Debye-Hückel formalism for the electrostatics, was used and found to qualitatively recapitulate the experimental results. Analysis of the energy landscapes of the designed versus wild-type proteins indicates the differences in refolding rates may be correlated with the degree of frustration of their respective energy landscapes. Our simulations indicate that naturally occurring wild-type proteins have frustrated folding landscapes due to the surface electrostatics. Optimization of the surface electrostatics seems to remove some of that frustration, leading to enhanced formation of native-like contacts in the transition-state ensembles (TSE) and providing a less frustrated energy landscape between the unfolded and TS ensembles. Macroscopically, this results in faster folding rates. Furthermore, analyses of pairwise distances and radii of gyration suggest that the less frustrated energy landscapes for optimized variants are a result of more compact unfolded and TS ensembles. These findings from our modeling demonstrates that this simple model may be used to: (i) gain a detailed understanding of charge-charge interactions and their effects on modulating the energy landscape of protein folding and (ii) qualitatively predict the kinetic behavior of protein surface electrostatic interactions.
Keywords: charge–charge interaction; computational design; energy landscape; protein folding; protein stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Electrostatics, structure prediction, and the energy landscapes for protein folding and binding.Protein Sci. 2016 Jan;25(1):255-69. doi: 10.1002/pro.2751. Epub 2015 Aug 8. Protein Sci. 2016. PMID: 26183799 Free PMC article.
-
Alterations of Nonconserved Residues Affect Protein Stability and Folding Dynamics through Charge-Charge Interactions.J Phys Chem B. 2015 Oct 15;119(41):13103-12. doi: 10.1021/acs.jpcb.5b08527. Epub 2015 Oct 2. J Phys Chem B. 2015. PMID: 26413861
-
Kinetic consequences of native state optimization of surface-exposed electrostatic interactions in the Fyn SH3 domain.Proteins. 2012 Mar;80(3):858-70. doi: 10.1002/prot.23243. Epub 2011 Dec 13. Proteins. 2012. PMID: 22161863
-
Linking computation and experiments to study the role of charge-charge interactions in protein folding and stability.Phys Biol. 2017 Feb 7;14(1):013002. doi: 10.1088/1478-3975/14/1/013002. Phys Biol. 2017. PMID: 28169222 Review.
-
Fuzziness and Frustration in the Energy Landscape of Protein Folding, Function, and Assembly.Acc Chem Res. 2021 Mar 2;54(5):1251-1259. doi: 10.1021/acs.accounts.0c00813. Epub 2021 Feb 8. Acc Chem Res. 2021. PMID: 33550810 Free PMC article. Review.
Cited by
-
Electrostatic control of calcineurin's intrinsically-disordered regulatory domain binding to calmodulin.Biochim Biophys Acta Gen Subj. 2018 Dec;1862(12):2651-2659. doi: 10.1016/j.bbagen.2018.07.027. Epub 2018 Jul 31. Biochim Biophys Acta Gen Subj. 2018. PMID: 30071273 Free PMC article.
-
The Role of Electrostatics and Folding Kinetics on the Thermostability of Homologous Cold Shock Proteins.J Chem Inf Model. 2020 Feb 24;60(2):546-561. doi: 10.1021/acs.jcim.9b00797. Epub 2020 Jan 17. J Chem Inf Model. 2020. PMID: 31910002 Free PMC article.
-
Effects of pH and Salt Concentration on Stability of a Protein G Variant Using Coarse-Grained Models.Biophys J. 2018 Jan 9;114(1):65-75. doi: 10.1016/j.bpj.2017.11.012. Biophys J. 2018. PMID: 29320697 Free PMC article.
-
Protein stability: computation, sequence statistics, and new experimental methods.Curr Opin Struct Biol. 2015 Aug;33:161-8. doi: 10.1016/j.sbi.2015.09.002. Curr Opin Struct Biol. 2015. PMID: 26497286 Free PMC article. Review.
-
The protonation state of an evolutionarily conserved histidine modulates domainswapping stability of FoxP1.Sci Rep. 2019 Apr 1;9(1):5441. doi: 10.1038/s41598-019-41819-5. Sci Rep. 2019. PMID: 30931977 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

