Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors
- PMID: 25564622
- PMCID: PMC4302835
- DOI: 10.1242/dev.116855
Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors
Abstract
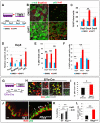
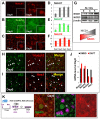
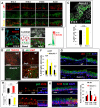
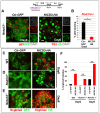
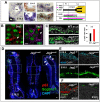
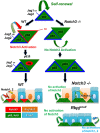
Basal cells are multipotent airway progenitors that generate distinct epithelial cell phenotypes crucial for homeostasis and repair of the conducting airways. Little is known about how these progenitor cells expand and transition to differentiation to form the pseudostratified airway epithelium in the developing and adult lung. Here, we show by genetic and pharmacological approaches that endogenous activation of Notch3 signaling selectively controls the pool of undifferentiated progenitors of upper airways available for differentiation. This mechanism depends on the availability of Jag1 and Jag2, and is key to generating a population of parabasal cells that later activates Notch1 and Notch2 for secretory-multiciliated cell fate selection. Disruption of this mechanism resulted in aberrant expansion of basal cells and altered pseudostratification. Analysis of human lungs showing similar abnormalities and decreased NOTCH3 expression in subjects with chronic obstructive pulmonary disease suggests an involvement of NOTCH3-dependent events in the pathogenesis of this condition.
Keywords: Airway differentiation; Basal cells; COPD; Jagged; Lung regeneration; Notch; Progenitor cells; p63.
© 2015. Published by The Company of Biologists Ltd.
Figures
Similar articles
-
Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung.Nature. 2015 Dec 3;528(7580):127-31. doi: 10.1038/nature15715. Epub 2015 Nov 18. Nature. 2015. PMID: 26580007
-
Notch signaling dynamics in the adult healthy prostate and in prostatic tumor development.Prostate. 2016 Jan;76(1):80-96. doi: 10.1002/pros.23102. Epub 2015 Sep 30. Prostate. 2016. PMID: 26419726
-
Distinct biological roles for the notch ligands Jagged-1 and Jagged-2.J Biol Chem. 2009 Jun 26;284(26):17766-74. doi: 10.1074/jbc.M109.003111. Epub 2009 Apr 27. J Biol Chem. 2009. PMID: 19398556 Free PMC article.
-
Cell and molecular biology of Notch.J Endocrinol. 2007 Sep;194(3):459-74. doi: 10.1677/JOE-07-0242. J Endocrinol. 2007. PMID: 17761886 Review.
-
Functional role of Notch signaling in the developing and postnatal heart.J Mol Cell Cardiol. 2008 Oct;45(4):495-504. doi: 10.1016/j.yjmcc.2008.02.273. Epub 2008 Mar 10. J Mol Cell Cardiol. 2008. PMID: 18410944 Review.
Cited by
-
The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future.Cell Stem Cell. 2020 Apr 2;26(4):482-502. doi: 10.1016/j.stem.2020.03.009. Cell Stem Cell. 2020. PMID: 32243808 Free PMC article. Review.
-
A novel method for expansion and differentiation of mouse tracheal epithelial cells in culture.Sci Rep. 2018 May 9;8(1):7349. doi: 10.1038/s41598-018-25799-6. Sci Rep. 2018. PMID: 29743551 Free PMC article.
-
Multi-resolution characterization of molecular taxonomies in bulk and single-cell transcriptomics data.Nucleic Acids Res. 2021 Sep 27;49(17):e98. doi: 10.1093/nar/gkab552. Nucleic Acids Res. 2021. PMID: 34226941 Free PMC article.
-
FGFR2 is required for airway basal cell self-renewal and terminal differentiation.Development. 2017 May 1;144(9):1600-1606. doi: 10.1242/dev.135681. Epub 2017 Mar 27. Development. 2017. PMID: 28348168 Free PMC article.
-
The emerging role of NOTCH3 receptor signalling in human lung diseases.Expert Rev Mol Med. 2022 Sep 2;24:e33. doi: 10.1017/erm.2022.27. Expert Rev Mol Med. 2022. PMID: 36052538 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

