Mechanism-based proteomic screening identifies targets of thioredoxin-like proteins
- PMID: 25561728
- PMCID: PMC4342480
- DOI: 10.1074/jbc.M114.597245
Mechanism-based proteomic screening identifies targets of thioredoxin-like proteins
Abstract
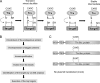
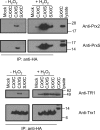
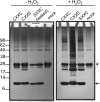
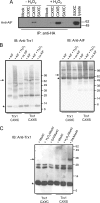
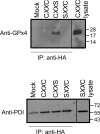
Thioredoxin (Trx)-fold proteins are protagonists of numerous cellular pathways that are subject to thiol-based redox control. The best characterized regulator of thiols in proteins is Trx1 itself, which together with thioredoxin reductase 1 (TR1) and peroxiredoxins (Prxs) comprises a key redox regulatory system in mammalian cells. However, there are numerous other Trx-like proteins, whose functions and redox interactors are unknown. It is also unclear if the principles of Trx1-based redox control apply to these proteins. Here, we employed a proteomic strategy to four Trx-like proteins containing CXXC motifs, namely Trx1, Rdx12, Trx-like protein 1 (Txnl1) and nucleoredoxin 1 (Nrx1), whose cellular targets were trapped in vivo using mutant Trx-like proteins, under conditions of low endogenous expression of these proteins. Prxs were detected as key redox targets of Trx1, but this approach also supported the detection of TR1, which is the Trx1 reductant, as well as mitochondrial intermembrane proteins AIF and Mia40. In addition, glutathione peroxidase 4 was found to be a Rdx12 redox target. In contrast, no redox targets of Txnl1 and Nrx1 could be detected, suggesting that their CXXC motifs do not engage in mixed disulfides with cellular proteins. For some Trx-like proteins, the method allowed distinguishing redox and non-redox interactions. Parallel, comparative analyses of multiple thiol oxidoreductases revealed differences in the functions of their CXXC motifs, providing important insights into thiol-based redox control of cellular processes.
Keywords: Mammal; Oxidation-reduction (Redox); Proteomics; Redox Regulation; Selenocysteine; Thiol; Thioredoxin; Thioredoxin Reductase.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells.Mol Plant. 2014 Jan;7(1):30-44. doi: 10.1093/mp/sst162. Epub 2013 Nov 19. Mol Plant. 2014. PMID: 24253198
-
Thioredoxin-1 redox signaling regulates cell survival in response to hyperoxia.Free Radic Biol Med. 2014 Oct;75:167-77. doi: 10.1016/j.freeradbiomed.2014.07.023. Epub 2014 Aug 6. Free Radic Biol Med. 2014. PMID: 25106706 Free PMC article.
-
Thioredoxin-dependent regulation of AIF-mediated DNA damage.Free Radic Biol Med. 2015 Oct;87:125-36. doi: 10.1016/j.freeradbiomed.2015.06.029. Epub 2015 Jun 25. Free Radic Biol Med. 2015. PMID: 26119781
-
The thioredoxin antioxidant system.Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review.
-
Thioredoxins and thioredoxin reductase in chloroplasts: A review.Gene. 2019 Jul 20;706:32-42. doi: 10.1016/j.gene.2019.04.041. Epub 2019 Apr 24. Gene. 2019. PMID: 31028868 Review.
Cited by
-
TXNL1 has dual functions as a redox active thioredoxin-like protein as well as an ATP- and redox-independent chaperone.Redox Biol. 2023 Nov;67:102897. doi: 10.1016/j.redox.2023.102897. Epub 2023 Sep 26. Redox Biol. 2023. PMID: 37804695 Free PMC article.
-
Differences in Reperfusion-Induced Mitochondrial Oxidative Stress and Cell Death Between Hippocampal CA1 and CA3 Subfields Are Due to the Mitochondrial Thioredoxin System.Antioxid Redox Signal. 2017 Sep 20;27(9):534-549. doi: 10.1089/ars.2016.6706. Epub 2017 Mar 7. Antioxid Redox Signal. 2017. PMID: 28129719 Free PMC article.
-
The Mia40/CHCHD4 Oxidative Folding System: Redox Regulation and Signaling in the Mitochondrial Intermembrane Space.Antioxidants (Basel). 2021 Apr 12;10(4):592. doi: 10.3390/antiox10040592. Antioxidants (Basel). 2021. PMID: 33921425 Free PMC article. Review.
-
Trapping redox partnerships in oxidant-sensitive proteins with a small, thiol-reactive cross-linker.Free Radic Biol Med. 2016 Dec;101:356-366. doi: 10.1016/j.freeradbiomed.2016.10.506. Epub 2016 Nov 2. Free Radic Biol Med. 2016. PMID: 27816612 Free PMC article.
-
Compensatory Protection of Thioredoxin-Deficient Cells from Etoposide-Induced Cell Death by Selenoprotein W via Interaction with 14-3-3.Int J Mol Sci. 2021 Sep 25;22(19):10338. doi: 10.3390/ijms221910338. Int J Mol Sci. 2021. PMID: 34638679 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

