Identifying cooperative transcription factors in yeast using multiple data sources
- PMID: 25559499
- PMCID: PMC4305981
- DOI: 10.1186/1752-0509-8-S5-S2
Identifying cooperative transcription factors in yeast using multiple data sources
Abstract
Background: Transcriptional regulation of gene expression is usually accomplished by multiple interactive transcription factors (TFs). Therefore, it is crucial to understand the precise cooperative interactions among TFs. Various kinds of experimental data including ChIP-chip, TF binding site (TFBS), gene expression, TF knockout and protein-protein interaction data have been used to identify cooperative TF pairs in existing methods. The nucleosome occupancy data is not yet used for this research topic despite that several researches have revealed the association between nucleosomes and TFBSs.
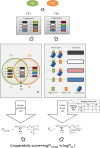
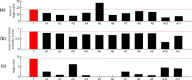
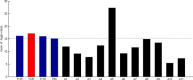
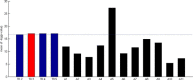
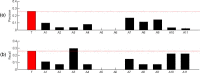
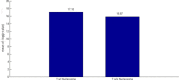
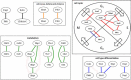
Results: In this study, we developed a novel method to infer the cooperativity between two TFs by integrating the TF-gene documented regulation, TFBS and nucleosome occupancy data. TF-gene documented regulation and TFBS data were used to determine the target genes of a TF, and the genome-wide nucleosome occupancy data was used to assess the nucleosome occupancy on TFBSs. Our method identifies cooperative TF pairs based on two biologically plausible assumptions. If two TFs cooperate, then (i) they should have a significantly higher number of common target genes than random expectation and (ii) their binding sites (in the promoters of their common target genes) should tend to be co-depleted of nucleosomes in order to make these binding sites simultaneously accessible to TF binding. Each TF pair is given a cooperativity score by our method. The higher the score is, the more likely a TF pair has cooperativity. Finally, a list of 27 cooperative TF pairs has been predicted by our method. Among these 27 TF pairs, 19 pairs are also predicted by existing methods. The other 8 pairs are novel cooperative TF pairs predicted by our method. The biological relevance of these 8 novel cooperative TF pairs is justified by the existence of protein-protein interactions and co-annotation in the same MIPS functional categories. Moreover, we adopted three performance indices to compare our predictions with 11 existing methods' predictions. We show that our method performs better than these 11 existing methods in identifying cooperative TF pairs in yeast. Finally, the cooperative TF network constructed from the 27 predicted cooperative TF pairs shows that our method has the power to find cooperative TF pairs of different biological processes.
Conclusion: Our method is effective in identifying cooperative TF pairs in yeast. Many of our predictions are validated by the literature, and our method outperforms 11 existing methods. We believe that our study will help biologists to understand the mechanisms of transcriptional regulation in eukaryotic cells.
Figures
Similar articles
-
Properly defining the targets of a transcription factor significantly improves the computational identification of cooperative transcription factor pairs in yeast.BMC Genomics. 2015;16 Suppl 12(Suppl 12):S10. doi: 10.1186/1471-2164-16-S12-S10. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26679776 Free PMC article.
-
A regulatory similarity measure using the location information of transcription factor binding sites in Saccharomyces cerevisiae.BMC Syst Biol. 2014;8 Suppl 5(Suppl 5):S9. doi: 10.1186/1752-0509-8-S5-S9. Epub 2014 Dec 12. BMC Syst Biol. 2014. PMID: 25560196 Free PMC article.
-
Detecting Cooperativity between Transcription Factors Based on Functional Coherence and Similarity of Their Target Gene Sets.PLoS One. 2016 Sep 13;11(9):e0162931. doi: 10.1371/journal.pone.0162931. eCollection 2016. PLoS One. 2016. PMID: 27623007 Free PMC article.
-
Building Transcription Factor Binding Site Models to Understand Gene Regulation in Plants.Mol Plant. 2019 Jun 3;12(6):743-763. doi: 10.1016/j.molp.2018.10.010. Epub 2018 Nov 15. Mol Plant. 2019. PMID: 30447332 Review.
-
A conserved role for transcription factor sumoylation in binding-site selection.Curr Genet. 2019 Dec;65(6):1307-1312. doi: 10.1007/s00294-019-00992-w. Epub 2019 May 15. Curr Genet. 2019. PMID: 31093693 Review.
Cited by
-
Removing Background Co-occurrences of Transcription Factor Binding Sites Greatly Improves the Prediction of Specific Transcription Factor Cooperations.Front Genet. 2018 May 29;9:189. doi: 10.3389/fgene.2018.00189. eCollection 2018. Front Genet. 2018. PMID: 29896218 Free PMC article.
-
Inference and interrogation of a coregulatory network in the context of lipid accumulation in Yarrowia lipolytica.NPJ Syst Biol Appl. 2017 Aug 11;3:21. doi: 10.1038/s41540-017-0024-1. eCollection 2017. NPJ Syst Biol Appl. 2017. PMID: 28955503 Free PMC article.
-
CoopTFD: a repository for predicted yeast cooperative transcription factor pairs.Database (Oxford). 2016 May 30;2016:baw092. doi: 10.1093/database/baw092. Print 2016. Database (Oxford). 2016. PMID: 27242036 Free PMC article.
-
YCRD: Yeast Combinatorial Regulation Database.PLoS One. 2016 Jul 8;11(7):e0159213. doi: 10.1371/journal.pone.0159213. eCollection 2016. PLoS One. 2016. PMID: 27392072 Free PMC article.
-
Properly defining the targets of a transcription factor significantly improves the computational identification of cooperative transcription factor pairs in yeast.BMC Genomics. 2015;16 Suppl 12(Suppl 12):S10. doi: 10.1186/1471-2164-16-S12-S10. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26679776 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

