Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage
- PMID: 25557546
- PMCID: PMC4416315
- DOI: 10.1016/j.molcel.2014.12.001
Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage
Abstract
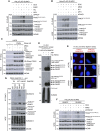
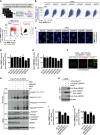
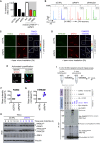
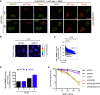
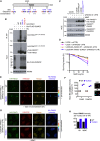
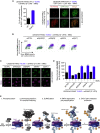
We show that central components of the Fanconi anemia (FA) DNA repair pathway, the tumor suppressor proteins FANCI and FANCD2 (the ID complex), are SUMOylated in response to replication fork stalling. The ID complex is SUMOylated in a manner that depends on the ATR kinase, the FA ubiquitin ligase core complex, and the SUMO E3 ligases PIAS1/PIAS4 and is antagonized by the SUMO protease SENP6. SUMOylation of the ID complex drives substrate selectivity by triggering its polyubiquitylation by the SUMO-targeted ubiquitin ligase RNF4 to promote its removal from sites of DNA damage via the DVC1-p97 ubiquitin segregase complex. Deregulation of ID complex SUMOylation compromises cell survival following replication stress. Our results uncover a regulatory role for SUMOylation in the FA pathway, and we propose that ubiquitin-SUMO signaling circuitry is a mechanism that contributes to the balance of activated ID complex dosage at sites of DNA damage.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Roles of the SUMO-related enzymes, PIAS1, PIAS4, and RNF4, in DNA double-strand break repair by homologous recombination.Biochem Biophys Res Commun. 2022 Feb 5;591:95-101. doi: 10.1016/j.bbrc.2021.12.099. Epub 2021 Dec 30. Biochem Biophys Res Commun. 2022. PMID: 35007836
-
c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4.Cell Cycle. 2015;14(12):1859-72. doi: 10.1080/15384101.2015.1040965. Cell Cycle. 2015. PMID: 25895136 Free PMC article.
-
Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays.Elife. 2020 Mar 13;9:e54128. doi: 10.7554/eLife.54128. Elife. 2020. PMID: 32167469 Free PMC article.
-
The Fanconi anemia ID2 complex: dueling saxes at the crossroads.Cell Cycle. 2014;13(19):2999-3015. doi: 10.4161/15384101.2014.956475. Cell Cycle. 2014. PMID: 25486561 Free PMC article. Review.
-
Regulation of the Fanconi Anemia DNA Repair Pathway by Phosphorylation and Monoubiquitination.Genes (Basel). 2021 Nov 5;12(11):1763. doi: 10.3390/genes12111763. Genes (Basel). 2021. PMID: 34828369 Free PMC article. Review.
Cited by
-
Stepwise phosphorylation and SUMOylation of PIDD1 drive PIDDosome assembly in response to DNA repair failure.Nat Commun. 2024 Oct 24;15(1):9195. doi: 10.1038/s41467-024-53412-0. Nat Commun. 2024. PMID: 39448602 Free PMC article.
-
Ring of Change: CDC48/p97 Drives Protein Dynamics at Chromatin.Front Genet. 2016 May 3;7:73. doi: 10.3389/fgene.2016.00073. eCollection 2016. Front Genet. 2016. PMID: 27200082 Free PMC article. Review.
-
Conditional degradation of SDE2 by the Arg/N-End rule pathway regulates stress response at replication forks.Nucleic Acids Res. 2019 May 7;47(8):3996-4010. doi: 10.1093/nar/gkz054. Nucleic Acids Res. 2019. PMID: 30698750 Free PMC article.
-
Fanconi anaemia and cancer: an intricate relationship.Nat Rev Cancer. 2018 Mar;18(3):168-185. doi: 10.1038/nrc.2017.116. Epub 2018 Jan 29. Nat Rev Cancer. 2018. PMID: 29376519 Review.
-
A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly.Nat Struct Mol Biol. 2015 Dec;22(12):959-67. doi: 10.1038/nsmb.3114. Epub 2015 Nov 2. Nat Struct Mol Biol. 2015. PMID: 26524493
References
-
- Adamo A., Collis S.J., Adelman C.A., Silva N., Horejsi Z., Ward J.D., Martinez-Perez E., Boulton S.J., La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 2010;39:25–35. - PubMed
-
- Akkari Y.M., Bateman R.L., Reifsteck C.A., D’Andrea A.D., Olson S.B., Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol. Genet. Metab. 2001;74:403–412. - PubMed
-
- Barysch S.V., Dittner C., Flotho A., Becker J., Melchior F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 2014;9:896–909. - PubMed
-
- Bergink S., Ammon T., Kern M., Schermelleh L., Leonhardt H., Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 2013;15:526–532. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

