Comparison of nested, multiplex, qPCR; FISH; SeptiFast and blood culture methods in detection and identification of bacteria and fungi in blood of patients with sepsis
- PMID: 25551203
- PMCID: PMC4302608
- DOI: 10.1186/s12866-014-0313-4
Comparison of nested, multiplex, qPCR; FISH; SeptiFast and blood culture methods in detection and identification of bacteria and fungi in blood of patients with sepsis
Abstract
Background: Microbiological diagnosis of sepsis relies primarily on blood culture data. This study compares four diagnostic methods, i.e. those developed by us: nested, multiplex, qPCR (qPCR) and FISH with commercial methods: SeptiFast (Roche) (SF) and BacT/ALERT® 3D blood culture system (bioMérieux). Blood samples were derived from adult patients with clinical symptoms of sepsis, according to SIRS criteria, hospitalized in the Intensive Care Unit.
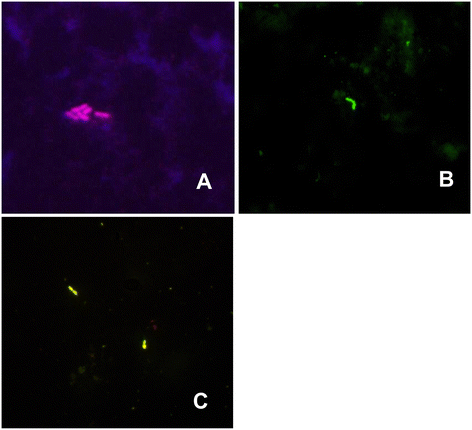
Results: Using qPCR, FISH, SF, and culture, microbial presence was found in 71.8%, 29.6%, 25.3%, and 36.6% of samples, respectively. It was demonstrated that qPCR was significantly more likely to detect microorganisms than the remaining methods; qPCR confirmed the results obtained with the SF kit in all cases wherein bacteria were detected with simultaneous confirmation of Gram-typing. All data collected through the FISH method were corroborated by qPCR.
Conclusions: The qPCR and FISH methods described in this study may constitute alternatives to blood culture and to the few existing commercial molecular assays since they enable the detection of the majority of microbial species, and the qPCR method allows their identification in a higher number of samples than the SF test. FISH made it possible to show the presence of microbes in a blood sample even before its culture.
Figures
Similar articles
-
Prospective evaluation of the SeptiFAST multiplex real-time PCR assay for surveillance and diagnosis of infections in haematological patients after allogeneic stem cell transplantation compared to routine microbiological assays and an in-house real-time PCR method.Mycoses. 2017 Dec;60(12):781-788. doi: 10.1111/myc.12662. Epub 2017 Sep 19. Mycoses. 2017. PMID: 28925082
-
Microbiological Diagnosis of Sepsis: Polymerase Chain Reaction System Versus Blood Cultures.Am J Crit Care. 2016 Jan;25(1):68-75. doi: 10.4037/ajcc2016728. Am J Crit Care. 2016. PMID: 26724297
-
A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood.BMC Microbiol. 2014 Jun 4;14:144. doi: 10.1186/1471-2180-14-144. BMC Microbiol. 2014. PMID: 24893651 Free PMC article.
-
Accuracy of LightCycler(®) SeptiFast for the detection and identification of pathogens in the blood of patients with suspected sepsis: a systematic review and meta-analysis.Intensive Care Med. 2015 Jan;41(1):21-33. doi: 10.1007/s00134-014-3553-8. Epub 2014 Nov 22. Intensive Care Med. 2015. PMID: 25416643 Review.
-
Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood.Clin Microbiol Infect. 2015 Apr;21(4):323-31. doi: 10.1016/j.cmi.2015.02.005. Epub 2015 Feb 14. Clin Microbiol Infect. 2015. PMID: 25686695 Review.
Cited by
-
Exploratory Investigation of Intestinal Function and Bacterial Translocation After Focal Cerebral Ischemia in the Mouse.Front Neurol. 2018 Nov 19;9:937. doi: 10.3389/fneur.2018.00937. eCollection 2018. Front Neurol. 2018. PMID: 30510535 Free PMC article.
-
Autophagy Strengthens Intestinal Mucosal Barrier by Attenuating Oxidative Stress in Severe Acute Pancreatitis.Dig Dis Sci. 2018 Apr;63(4):910-919. doi: 10.1007/s10620-018-4962-2. Epub 2018 Feb 9. Dig Dis Sci. 2018. PMID: 29427225
-
Intra-abdominal Candida spp infection in acute abdomen in a quality assurance (QA)-certified academic setting.J Clin Pathol. 2017 Jul;70(7):579-583. doi: 10.1136/jclinpath-2016-203936. Epub 2016 Dec 9. J Clin Pathol. 2017. PMID: 27941028 Free PMC article.
-
Optimization and Testing of a Commercial Viability PCR Protocol to Detect Escherichia coli in Whole Blood.Microorganisms. 2024 Apr 10;12(4):765. doi: 10.3390/microorganisms12040765. Microorganisms. 2024. PMID: 38674709 Free PMC article.
-
Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia.Eur J Clin Microbiol Infect Dis. 2017 Feb;36(2):329-336. doi: 10.1007/s10096-016-2805-7. Epub 2016 Oct 22. Eur J Clin Microbiol Infect Dis. 2017. PMID: 27771780 Free PMC article.
References
-
- Burdino E, Ruggiero T, Allice T, Milia MG, Gregori G, Milano R, Cerutti F, De Rosa FG, Manno E, Caramello P, Di Perri G, Ghisetti V. Combination of conventional blood cultures and the SeptiFast molecular test in patients with suspected sepsis for the identification of bloodstream pathogens. Diagn Microbiol Infect Dis. 2014;79:287–292. doi: 10.1016/j.diagmicrobio.2014.03.018. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical