Dimerization of cAMP phosphodiesterase-4 (PDE4) in living cells requires interfaces located in both the UCR1 and catalytic unit domains
- PMID: 25546709
- PMCID: PMC4371794
- DOI: 10.1016/j.cellsig.2014.12.009
Dimerization of cAMP phosphodiesterase-4 (PDE4) in living cells requires interfaces located in both the UCR1 and catalytic unit domains
Abstract
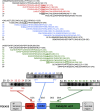
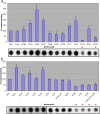
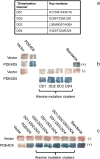
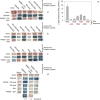
PDE4 family cAMP phosphodiesterases play a pivotal role in determining compartmentalised cAMP signalling through targeted cAMP breakdown. Expressing the widely found PDE4D5 isoform, as both bait and prey in a yeast 2-hybrid system, we demonstrated interaction consistent with the notion that long PDE4 isoforms form dimers. Four potential dimerization sites were uncovered using a scanning peptide array approach, where a recombinant purified PDE4D5 fusion protein was used to probe a 25-mer library of overlapping peptides covering the entire PDE4D5 sequence. Key residues involved in PDE4D5 dimerization were defined using a site-directed mutagenesis programme directed by an alanine scanning peptide array approach. Critical residues stabilising PDE4D5 dimerization were defined within the regulatory UCR1 region found in long, but not short, PDE4 isoforms, namely the Arg(173), Asn(174) and Asn(175) (DD1) cluster. Disruption of the DD1 cluster was not sufficient, in itself, to destabilise PDE4D5 homodimers. Instead, disruption of an additional interface, located on the PDE4 catalytic unit, was also required to convert PDE4D5 into a monomeric form. This second dimerization site on the conserved PDE4 catalytic unit is dependent upon a critical ion pair interaction. This involves Asp(463) and Arg(499) in PDE4D5, which interact in a trans fashion involving the two PDE4D5 molecules participating in the homodimer. PDE4 long isoforms adopt a dimeric state in living cells that is underpinned by two key contributory interactions, one involving the UCR modules and one involving an interface on the core catalytic domain. We propose that short forms do not adopt a dimeric configuration because, in the absence of the UCR1 module, residual engagement of the remaining core catalytic domain interface provides insufficient free energy to drive dimerization. The functioning of PDE4 long and short forms is thus poised to be inherently distinct due to this difference in quaternary structure.
Keywords: Dimerisation; Dimerization; PDE4; Phosphodiesterase; Rolipram; cAMP; cyclic AMP.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Identification of a multifunctional docking site on the catalytic unit of phosphodiesterase-4 (PDE4) that is utilised by multiple interaction partners.Biochem J. 2017 Feb 15;474(4):597-609. doi: 10.1042/BCJ20160849. Epub 2016 Dec 19. Biochem J. 2017. PMID: 27993970 Free PMC article.
-
Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1).Br J Pharmacol. 2002 Jun;136(3):421-33. doi: 10.1038/sj.bjp.0704743. Br J Pharmacol. 2002. PMID: 12023945 Free PMC article.
-
Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through Protein Kinase A (PKA).Cell Signal. 2008 Dec;20(12):2356-69. doi: 10.1016/j.cellsig.2008.09.017. Epub 2008 Oct 2. Cell Signal. 2008. PMID: 18845247
-
PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization.Biochem J. 2003 Feb 15;370(Pt 1):1-18. doi: 10.1042/BJ20021698. Biochem J. 2003. PMID: 12444918 Free PMC article. Review.
-
The role of ERK2 docking and phosphorylation of PDE4 cAMP phosphodiesterase isoforms in mediating cross-talk between the cAMP and ERK signalling pathways.Biochem Soc Trans. 2003 Dec;31(Pt 6):1186-90. doi: 10.1042/bst0311186. Biochem Soc Trans. 2003. PMID: 14641023 Review.
Cited by
-
Small-molecule allosteric activators of PDE4 long form cyclic AMP phosphodiesterases.Proc Natl Acad Sci U S A. 2019 Jul 2;116(27):13320-13329. doi: 10.1073/pnas.1822113116. Epub 2019 Jun 17. Proc Natl Acad Sci U S A. 2019. PMID: 31209056 Free PMC article.
-
Altered phosphorylation, electrophysiology, and behavior on attenuation of PDE4B action in hippocampus.BMC Neurosci. 2017 Dec 2;18(1):77. doi: 10.1186/s12868-017-0396-6. BMC Neurosci. 2017. PMID: 29197324 Free PMC article.
-
Dominant-Negative Attenuation of cAMP-Selective Phosphodiesterase PDE4D Action Affects Learning and Behavior.Int J Mol Sci. 2020 Aug 9;21(16):5704. doi: 10.3390/ijms21165704. Int J Mol Sci. 2020. PMID: 32784895 Free PMC article.
-
Phenotypic, chemical and functional characterization of cyclic nucleotide phosphodiesterase 4 (PDE4) as a potential anthelmintic drug target.PLoS Negl Trop Dis. 2017 Jul 13;11(7):e0005680. doi: 10.1371/journal.pntd.0005680. eCollection 2017 Jul. PLoS Negl Trop Dis. 2017. PMID: 28704396 Free PMC article.
-
The analgesic effect of rolipram is associated with the inhibition of the activation of the spinal astrocytic JNK/CCL2 pathway in bone cancer pain.Int J Mol Med. 2016 Nov;38(5):1433-1442. doi: 10.3892/ijmm.2016.2763. Epub 2016 Sep 30. Int J Mol Med. 2016. PMID: 28025994 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

