Quantitative shearing linear amplification polymerase chain reaction: an improved method for quantifying lentiviral vector insertion sites in transplanted hematopoietic cell systems
- PMID: 25545666
- PMCID: PMC4337463
- DOI: 10.1089/hgtb.2014.122
Quantitative shearing linear amplification polymerase chain reaction: an improved method for quantifying lentiviral vector insertion sites in transplanted hematopoietic cell systems
Abstract
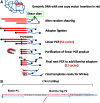
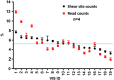
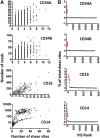
In gene therapy trials targeting blood disorders, it is important to detect dominance of transduced hematopoietic stem cell (HSC) clones arising from vector insertion site (VIS) effects. Current methods for VIS analysis often do not have defined levels of quantitative accuracy and therefore can fail to detect early clonal dominance. We have developed a rapid and inexpensive method for measuring clone size based on random shearing of genomic DNA, minimal exponential PCR amplification, and shear site counts as a quantitative endpoint. This quantitative shearing linear amplification PCR (qsLAM PCR) assay utilizes an internal control sample containing 19 lentiviral insertion sites per cell that is mixed with polyclonal samples derived from transduced human CD34+ cells. Samples were analyzed from transplanted pigtail macaques and from a participant in our X-linked severe combined immunodeficiency (XSCID) lentiviral vector trial and yielded controlled and quantitative results in all cases. One case of early clonal dominance was detected in a monkey transplanted with limiting numbers of transduced HSCs, while the clinical samples from the XSCID trial participant showed highly diverse clonal representation. These studies demonstrate that qsLAM PCR is a facile and quantitative assay for measuring clonal repertoires in subjects enrolled in human gene therapy trials using lentiviral-transduced HSCs.
Figures
Similar articles
-
Comparative clonal analysis of reconstitution kinetics after transplantation of hematopoietic stem cells gene marked with a lentiviral SIN or a γ-retroviral LTR vector.Exp Hematol. 2013 Jan;41(1):28-38.e3. doi: 10.1016/j.exphem.2012.09.003. Epub 2012 Sep 16. Exp Hematol. 2013. PMID: 22989760
-
Lentiviral-Mediated Gene Therapy in Fanconi Anemia-A Mice Reveals Long-Term Engraftment and Continuous Turnover of Corrected HSCs.Curr Gene Ther. 2015;15(6):550-62. doi: 10.2174/1566523215666150929110903. Curr Gene Ther. 2015. PMID: 26415575
-
Safety and efficacy of a tCD25 preselective combination anti-HIV lentiviral vector in human hematopoietic stem and progenitor cells.Stem Cells. 2015 Mar;33(3):870-9. doi: 10.1002/stem.1919. Stem Cells. 2015. PMID: 25524029
-
Hematopoietic stem cell gene transfer for the treatment of hemoglobin disorders.Hematology Am Soc Hematol Educ Program. 2009:690-7. doi: 10.1182/asheducation-2009.1.690. Hematology Am Soc Hematol Educ Program. 2009. PMID: 20008255 Review.
-
Gene therapy studies in a canine model of X-linked severe combined immunodeficiency.Hum Gene Ther Clin Dev. 2015 Mar;26(1):50-6. doi: 10.1089/humc.2015.004. Epub 2015 Feb 24. Hum Gene Ther Clin Dev. 2015. PMID: 25603151 Free PMC article. Review.
Cited by
-
Evidence for the in vivo safety of insulated foamy viral vectors.Gene Ther. 2017 Mar;24(3):187-198. doi: 10.1038/gt.2016.88. Epub 2016 Dec 26. Gene Ther. 2017. PMID: 28024082 Free PMC article.
-
Prostaglandin E2 Increases Lentiviral Vector Transduction Efficiency of Adult Human Hematopoietic Stem and Progenitor Cells.Mol Ther. 2018 Jan 3;26(1):320-328. doi: 10.1016/j.ymthe.2017.09.025. Epub 2017 Oct 5. Mol Ther. 2018. PMID: 29102562 Free PMC article.
-
An analytical pipeline for identifying and mapping the integration sites of HIV and other retroviruses.BMC Genomics. 2020 Mar 9;21(1):216. doi: 10.1186/s12864-020-6647-4. BMC Genomics. 2020. PMID: 32151239 Free PMC article.
-
Lentiviral Gene Therapy Combined with Low-Dose Busulfan in Infants with SCID-X1.N Engl J Med. 2019 Apr 18;380(16):1525-1534. doi: 10.1056/NEJMoa1815408. N Engl J Med. 2019. PMID: 30995372 Free PMC article. Clinical Trial.
-
Integrome signatures of lentiviral gene therapy for SCID-X1 patients.Sci Adv. 2023 Oct 6;9(40):eadg9959. doi: 10.1126/sciadv.adg9959. Epub 2023 Oct 6. Sci Adv. 2023. PMID: 37801507 Free PMC article.
References
-
- Dropulic B. Lentiviral vectors: their molecular design, safety, and use in laboratory and preclinical research. Hum Gene Ther 2011;22:649–657 - PubMed
-
- Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther 2006;13:1031–1049 - PubMed
-
- Braun CJ, Boztug K, Paruzynski A, et al. . Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014;6:227ra33 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

