Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution
- PMID: 25534808
- PMCID: PMC5898198
- DOI: 10.1038/nrmicro3389
Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution
Abstract
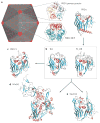
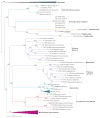
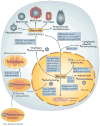
Polintons (also known as Mavericks) are large DNA transposons that are widespread in the genomes of eukaryotes. We have recently shown that Polintons encode virus capsid proteins, which suggests that these transposons might form virions, at least under some conditions. In this Opinion article, we delineate the evolutionary relationships among bacterial tectiviruses, Polintons, adenoviruses, virophages, large and giant DNA viruses of eukaryotes of the proposed order 'Megavirales', and linear mitochondrial and cytoplasmic plasmids. We hypothesize that Polintons were the first group of eukaryotic double-stranded DNA viruses to evolve from bacteriophages and that they gave rise to most large DNA viruses of eukaryotes and various other selfish genetic elements.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses.Ann N Y Acad Sci. 2015 Apr;1341(1):10-24. doi: 10.1111/nyas.12728. Epub 2015 Feb 27. Ann N Y Acad Sci. 2015. PMID: 25727355 Free PMC article. Review.
-
Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems.Curr Opin Microbiol. 2016 Jun;31:25-33. doi: 10.1016/j.mib.2016.01.006. Epub 2016 Feb 1. Curr Opin Microbiol. 2016. PMID: 26836982 Free PMC article. Review.
-
A novel group of diverse Polinton-like viruses discovered by metagenome analysis.BMC Biol. 2015 Nov 11;13:95. doi: 10.1186/s12915-015-0207-4. BMC Biol. 2015. PMID: 26560305 Free PMC article.
-
Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity.Curr Opin Virol. 2017 Aug;25:7-15. doi: 10.1016/j.coviro.2017.06.008. Epub 2017 Jun 30. Curr Opin Virol. 2017. PMID: 28672161 Free PMC article. Review.
-
Disentangling the origins of virophages and polintons.Curr Opin Virol. 2017 Aug;25:59-65. doi: 10.1016/j.coviro.2017.07.011. Epub 2017 Aug 9. Curr Opin Virol. 2017. PMID: 28802203 Review.
Cited by
-
Deep landscape update of dispersed and tandem repeats in the genome model of the red jungle fowl, Gallus gallus, using a series of de novo investigating tools.BMC Genomics. 2016 Aug 19;17(1):659. doi: 10.1186/s12864-016-3015-5. BMC Genomics. 2016. PMID: 27542599 Free PMC article.
-
Viral dUTPases: Modulators of Innate Immunity.Biomolecules. 2022 Jan 28;12(2):227. doi: 10.3390/biom12020227. Biomolecules. 2022. PMID: 35204728 Free PMC article. Review.
-
Unraveling Protein Interactions between the Temperate Virus Bam35 and Its Bacillus Host Using an Integrative Yeast Two Hybrid-High Throughput Sequencing Approach.Int J Mol Sci. 2021 Oct 14;22(20):11105. doi: 10.3390/ijms222011105. Int J Mol Sci. 2021. PMID: 34681765 Free PMC article.
-
Cellular homologs of the double jelly-roll major capsid proteins clarify the origins of an ancient virus kingdom.Proc Natl Acad Sci U S A. 2022 Feb 1;119(5):e2120620119. doi: 10.1073/pnas.2120620119. Proc Natl Acad Sci U S A. 2022. PMID: 35078938 Free PMC article.
-
Bacteriophage GC1, a Novel Tectivirus Infecting Gluconobacter Cerinus, an Acetic Acid Bacterium Associated with Wine-Making.Viruses. 2018 Jan 16;10(1):39. doi: 10.3390/v10010039. Viruses. 2018. PMID: 29337868 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

