The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses
- PMID: 25520508
- PMCID: PMC4325707
- DOI: 10.1128/JVI.02791-14
The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses
Abstract
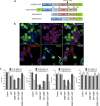
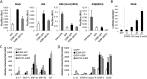
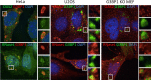
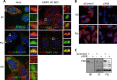
Stress granules (SGs) are cytoplasmic storage sites containing translationally silenced mRNPs that can be released to resume translation after stress subsides. We previously showed that poliovirus 3C proteinase cleaves the SG-nucleating protein G3BP1, blocking the ability of cells to form SGs late in infection. Many other viruses also target G3BP1 and inhibit SG formation, but the reasons why these functions evolved are unclear. Previously, we also showed a link between G3BP1-induced SGs and protein kinase R (PKR)-mediated translational control, but the mechanism of PKR interplay with SG and the antiviral consequences are unknown. Here, we show that G3BP1 exhibits antiviral activity against several enteroviruses, whereas truncated G3BP1 that cannot form SGs does not. G3BP1-induced SGs are linked to activation of innate immune transcriptional responses through NF-κB and JNK. The G3BP1-induced SGs also recruit PKR and other antiviral proteins. We show that the PXXP domain within G3BP1 is essential for the recruitment of PKR to SGs, for eIF2α phosphorylation driven by PKR, and for nucleating SGs of normal composition. We also show that deletion of the PXXP domain in G3BP1 compromises its antiviral activity. These findings tie PKR activation to its recruitment to SGs by G3BP1 and indicate that G3BP1 promotes innate immune responses at both the transcriptional and translational levels and integrates cellular stress responses and innate immunity.
Importance: Stress granules appear during virus infection, and their importance is not well understood. Previously, it was assumed that they were nonfunctional artifacts associated with cellular stress. PKR is a well-known antiviral protein; however, its regulation in cells is not well understood. Our work links cellular stress granules with activation of PKR and other innate immune pathways through the activity of G3BP1, a critical stress granule component. The ability of stress granules and G3BP1 to activate PKR and other innate immune transcriptional responses indicates that G3BP1 is an antiviral protein. This work helps to refine a longstanding paradigm indicating stress granules are inert structures and explains why G3BP1 is subverted by many viruses to promote a productive infection.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1.mBio. 2015 Mar 17;6(2):e02486. doi: 10.1128/mBio.02486-14. mBio. 2015. PMID: 25784705 Free PMC article.
-
Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article.
-
TDRD3 is an antiviral restriction factor that promotes IFN signaling with G3BP1.PLoS Pathog. 2022 Jan 27;18(1):e1010249. doi: 10.1371/journal.ppat.1010249. eCollection 2022 Jan. PLoS Pathog. 2022. PMID: 35085371 Free PMC article.
-
A closer look at mammalian antiviral condensates.Biochem Soc Trans. 2024 Jun 26;52(3):1393-1404. doi: 10.1042/BST20231296. Biochem Soc Trans. 2024. PMID: 38778761 Free PMC article. Review.
-
Pro-Viral and Anti-Viral Roles of the RNA-Binding Protein G3BP1.Viruses. 2023 Feb 6;15(2):449. doi: 10.3390/v15020449. Viruses. 2023. PMID: 36851663 Free PMC article. Review.
Cited by
-
A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates.FASEB J. 2020 Aug;34(8):9832-9842. doi: 10.1096/fj.202001351. Epub 2020 Jun 20. FASEB J. 2020. PMID: 32562316 Free PMC article.
-
Novel stress granule-like structures are induced via a paracrine mechanism during viral infection.J Cell Sci. 2022 Feb 15;135(4):jcs259194. doi: 10.1242/jcs.259194. Epub 2022 Feb 24. J Cell Sci. 2022. PMID: 35098996 Free PMC article.
-
Differences between acute and chronic stress granules, and how these differences may impact function in human disease.Biochem Pharmacol. 2019 Apr;162:123-131. doi: 10.1016/j.bcp.2018.10.009. Epub 2018 Oct 14. Biochem Pharmacol. 2019. PMID: 30326201 Free PMC article. Review.
-
Essential Role of Enterovirus 2A Protease in Counteracting Stress Granule Formation and the Induction of Type I Interferon.J Virol. 2019 May 1;93(10):e00222-19. doi: 10.1128/JVI.00222-19. Print 2019 May 15. J Virol. 2019. PMID: 30867299 Free PMC article.
-
Defining the Role of Stress Granules in Innate Immune Suppression by the Herpes Simplex Virus 1 Endoribonuclease VHS.J Virol. 2018 Jul 17;92(15):e00829-18. doi: 10.1128/JVI.00829-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793959 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

