Role of membrane contact sites in protein import into mitochondria
- PMID: 25514890
- PMCID: PMC4353355
- DOI: 10.1002/pro.2625
Role of membrane contact sites in protein import into mitochondria
Abstract
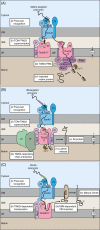
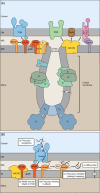
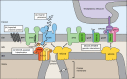
Mitochondria import more than 1,000 different proteins from the cytosol. The proteins are synthesized as precursors on cytosolic ribosomes and are translocated by protein transport machineries of the mitochondrial membranes. Five main pathways for protein import into mitochondria have been identified. Most pathways use the translocase of the outer mitochondrial membrane (TOM) as the entry gate into mitochondria. Depending on specific signals contained in the precursors, the proteins are subsequently transferred to different intramitochondrial translocases. In this article, we discuss the connection between protein import and mitochondrial membrane architecture. Mitochondria possess two membranes. It is a long-standing question how contact sites between outer and inner membranes are formed and which role the contact sites play in the translocation of precursor proteins. A major translocation contact site is formed between the TOM complex and the presequence translocase of the inner membrane (TIM23 complex), promoting transfer of presequence-carrying preproteins to the mitochondrial inner membrane and matrix. Recent findings led to the identification of contact sites that involve the mitochondrial contact site and cristae organizing system (MICOS) of the inner membrane. MICOS plays a dual role. It is crucial for maintaining the inner membrane cristae architecture and forms contacts sites to the outer membrane that promote translocation of precursor proteins into the intermembrane space and outer membrane of mitochondria. The view is emerging that the mitochondrial protein translocases do not function as independent units, but are embedded in a network of interactions with machineries that control mitochondrial activity and architecture.
Keywords: MICOS; contact site; membrane architecture; mitochondria; protein sorting; protein translocase.
© 2014 The Protein Society.
Figures

Similar articles
-
A MICOS-TIM22 Association Promotes Carrier Import into Human Mitochondria.J Mol Biol. 2019 Jul 12;431(15):2835-2851. doi: 10.1016/j.jmb.2019.05.015. Epub 2019 May 17. J Mol Biol. 2019. PMID: 31103774
-
Mitochondrial preprotein translocases as dynamic molecular machines.FEMS Yeast Res. 2006 Sep;6(6):849-61. doi: 10.1111/j.1567-1364.2006.00134.x. FEMS Yeast Res. 2006. PMID: 16911507 Review.
-
Unlocking the presequence import pathway.Trends Cell Biol. 2015 May;25(5):265-75. doi: 10.1016/j.tcb.2014.12.001. Epub 2014 Dec 23. Trends Cell Biol. 2015. PMID: 25542066 Review.
-
Cooperation of translocase complexes in mitochondrial protein import.J Cell Biol. 2007 Nov 19;179(4):585-91. doi: 10.1083/jcb.200708199. Epub 2007 Nov 12. J Cell Biol. 2007. PMID: 17998403 Free PMC article. Review.
-
Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria.J Mol Biol. 2015 Mar 13;427(5):1075-84. doi: 10.1016/j.jmb.2014.07.015. Epub 2014 Jul 30. J Mol Biol. 2015. PMID: 25083920
Cited by
-
SARS-CoV-2 and other coronaviruses negatively influence mitochondrial quality control: beneficial effects of melatonin.Pharmacol Ther. 2021 Aug;224:107825. doi: 10.1016/j.pharmthera.2021.107825. Epub 2021 Mar 1. Pharmacol Ther. 2021. PMID: 33662449 Free PMC article. Review.
-
Mitochondria as intracellular signaling platforms in health and disease.J Cell Biol. 2020 May 4;219(5):e202002179. doi: 10.1083/jcb.202002179. J Cell Biol. 2020. PMID: 32320464 Free PMC article. Review.
-
Unbiased Mitoproteome Analyses Confirm Non-canonical RNA, Expanded Codon Translations.Comput Struct Biotechnol J. 2016 Oct 5;14:391-403. doi: 10.1016/j.csbj.2016.09.004. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27830053 Free PMC article.
-
MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria.J Cell Biol. 2016 Jun 6;213(5):525-34. doi: 10.1083/jcb.201602007. Epub 2016 May 30. J Cell Biol. 2016. PMID: 27241913 Free PMC article.
-
The Diversity of the Mitochondrial Outer Membrane Protein Import Channels: Emerging Targets for Modulation.Molecules. 2021 Jul 4;26(13):4087. doi: 10.3390/molecules26134087. Molecules. 2021. PMID: 34279427 Free PMC article. Review.
References
-
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. - PubMed
-
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. - PubMed
-
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

