Mechanistic insights into xanthine oxidoreductase from development studies of candidate drugs to treat hyperuricemia and gout
- PMID: 25501928
- PMCID: PMC4334109
- DOI: 10.1007/s00775-014-1210-x
Mechanistic insights into xanthine oxidoreductase from development studies of candidate drugs to treat hyperuricemia and gout
Abstract
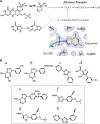
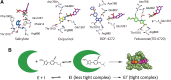
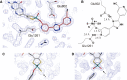
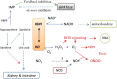
Xanthine oxidoreductase (XOR), which is widely distributed from humans to bacteria, has a key role in purine catabolism, catalyzing two steps of sequential hydroxylation from hypoxanthine to xanthine and from xanthine to urate at its molybdenum cofactor (Moco). Human XOR is considered to be a target of drugs not only for therapy of hyperuricemia and gout, but also potentially for a wide variety of other diseases. In this review, we focus on studies of XOR inhibitors and their implications for understanding the chemical nature and reaction mechanism of the Moco active site of XOR. We also discuss further experimental or clinical studies that would be helpful to clarify remaining issues.
Figures



Similar articles
-
Chemical nature and reaction mechanisms of the molybdenum cofactor of xanthine oxidoreductase.Curr Pharm Des. 2013;19(14):2606-14. doi: 10.2174/1381612811319140010. Curr Pharm Des. 2013. PMID: 23116398 Free PMC article. Review.
-
Investigation of the Inhibition Mechanism of Xanthine Oxidoreductase by Oxipurinol: A Computational Study.J Chem Inf Model. 2023 Jul 10;63(13):4190-4206. doi: 10.1021/acs.jcim.3c00624. Epub 2023 Jun 15. J Chem Inf Model. 2023. PMID: 37319436 Free PMC article.
-
Hyperuricemia-Related Diseases and Xanthine Oxidoreductase (XOR) Inhibitors: An Overview.Med Sci Monit. 2016 Jul 17;22:2501-12. doi: 10.12659/msm.899852. Med Sci Monit. 2016. PMID: 27423335 Free PMC article. Review.
-
Discriminating Susceptibility of Xanthine Oxidoreductase Family to Metals.Microbiol Spectr. 2023 Aug 17;11(4):e0481422. doi: 10.1128/spectrum.04814-22. Epub 2023 Jul 17. Microbiol Spectr. 2023. PMID: 37458582 Free PMC article.
-
Xanthine oxidoreductase and its inhibitors: relevance for gout.Clin Sci (Lond). 2016 Dec 1;130(23):2167-2180. doi: 10.1042/CS20160010. Clin Sci (Lond). 2016. PMID: 27798228 Review.
Cited by
-
Purine Metabolism Dysfunctions: Experimental Methods of Detection and Diagnostic Potential.Int J Mol Sci. 2023 Apr 10;24(8):7027. doi: 10.3390/ijms24087027. Int J Mol Sci. 2023. PMID: 37108190 Free PMC article. Review.
-
A pharmacokinetic-pharmacodynamic study of a single dose of febuxostat in healthy subjects.Br J Clin Pharmacol. 2020 Dec;86(12):2486-2496. doi: 10.1111/bcp.14357. Epub 2020 Jun 18. Br J Clin Pharmacol. 2020. PMID: 32386239 Free PMC article.
-
Immune Modulation by Myeloid-Derived Suppressor Cells in Diabetic Kidney Disease.Int J Mol Sci. 2022 Oct 31;23(21):13263. doi: 10.3390/ijms232113263. Int J Mol Sci. 2022. PMID: 36362050 Free PMC article. Review.
-
The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.FEBS J. 2015 Aug;282(16):3075-90. doi: 10.1111/febs.13277. Epub 2015 Apr 13. FEBS J. 2015. PMID: 25817260 Free PMC article.
-
Metabolomic Characteristics of Liver and Cecum Contents in High-Fat-Diet-Induced Obese Mice Intervened with Lactobacillus plantarum FRT10.Foods. 2022 Aug 18;11(16):2491. doi: 10.3390/foods11162491. Foods. 2022. PMID: 36010491 Free PMC article.
References
-
- Hille R, Nishino T. FASEB J. 1995;9:995–1003. - PubMed
-
- Schardinger F (1902) Untersuch Nahrungs Genussmittel, pp 1113–1121
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

