A knockdown with smoke model reveals FHIT as a repressor of Heme oxygenase 1
- PMID: 25486479
- PMCID: PMC4614990
- DOI: 10.4161/15384101.2014.946858
A knockdown with smoke model reveals FHIT as a repressor of Heme oxygenase 1
Abstract
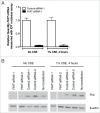
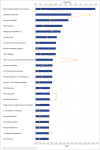
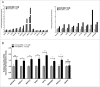
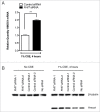
Fragile histidine triad (FHIT) gene deletions are among the earliest and most frequent events in carcinogenesis, particularly in carcinogen-exposed tissues. Though FHIT has been established as an authentic tumor suppressor, the mechanism underlying tumor suppression remains opaque. Most experiments designed to clarify FHIT function have analyzed the consequence of re-expressing FHIT in FHIT-negative cells. However, carcinogenesis occurs in cells that transition from FHIT-positive to FHIT-negative. To better understand cancer development, we induced FHIT loss in human bronchial epithelial cells with RNA interference. Because FHIT is a demonstrated target of carcinogens in cigarette smoke, we combined FHIT silencing with cigarette smoke extract (CSE) exposure and measured gene expression consequences by RNA microarray. The data indicate that FHIT loss enhances the expression of a set of oxidative stress response genes after exposure to CSE, including the cytoprotective enzyme heme oxygenase 1 (HMOX1) at the RNA and protein levels. Data are consistent with a mechanism in which Fhit protein is required for accumulation of the transcriptional repressor of HMOX1, Bach1 protein. We posit that by allowing superinduction of oxidative stress response genes, loss of FHIT creates a survival advantage that promotes carcinogenesis.
Keywords: ARE, antioxidant response element; ApppA, diadenosine triphosphate; BACH1; BACH1, BTB and CNC homology 1 gene; BMC, bone marrow cell; CPT, camptothecin; CSE, cigarette smoke extract; Cigarette smoke; FHIT; FHIT, fragile histidine triad gene; HMOX1; HMOX1, heme oxygenase 1 gene; MMC, mitomycin C; NRF2; Nrf2, nuclear factor erythroid derived 2-like 2 protein; Oxidative Stress; RNAi, RNA interference; ROS, reactive oxygen species; qRT-PCR, quantitative real time PCR; siRNA, short interfering RNA.
Figures




Similar articles
-
Prolonged cigarette smoke exposure decreases heme oxygenase-1 and alters Nrf2 and Bach1 expression in human macrophages: roles of the MAP kinases ERK(1/2) and JNK.FEBS Lett. 2009 Nov 3;583(21):3508-18. doi: 10.1016/j.febslet.2009.10.010. Epub 2009 Oct 12. FEBS Lett. 2009. PMID: 19822148
-
Dual regulation of skin sensitizer-induced HMOX1 expression by Bach1 and Nrf2: Comparison to regulation of the AKR1C2-ARE element in the KeratinoSens cell line.Toxicol Appl Pharmacol. 2015 Nov 1;288(3):281-8. doi: 10.1016/j.taap.2015.07.027. Epub 2015 Aug 2. Toxicol Appl Pharmacol. 2015. PMID: 26244607
-
Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1.Nucleic Acids Res. 2007;35(21):7074-86. doi: 10.1093/nar/gkm638. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17942419 Free PMC article.
-
Silencing Bach1 alters aging-related changes in the expression of Nrf2-regulated genes in primary human bronchial epithelial cells.Arch Biochem Biophys. 2019 Sep 15;672:108074. doi: 10.1016/j.abb.2019.108074. Epub 2019 Aug 15. Arch Biochem Biophys. 2019. PMID: 31422075 Review.
-
The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation.Antioxid Redox Signal. 2006 Jan-Feb;8(1-2):107-18. doi: 10.1089/ars.2006.8.107. Antioxid Redox Signal. 2006. PMID: 16487043 Review.
Cited by
-
Fhit-Fdxr interaction in the mitochondria: modulation of reactive oxygen species generation and apoptosis in cancer cells.Cell Death Dis. 2019 Feb 15;10(3):147. doi: 10.1038/s41419-019-1414-7. Cell Death Dis. 2019. PMID: 30770797 Free PMC article.
-
Heme oxygenase-1 gene promoter polymorphisms are associated with coronary heart disease and restenosis after percutaneous coronary intervention: a meta-analysis.Oncotarget. 2016 Dec 13;7(50):83437-83450. doi: 10.18632/oncotarget.13118. Oncotarget. 2016. PMID: 27825138 Free PMC article. Review.
-
Cancer-associated fibroblasts promote cancer cell growth through a miR-7-RASSF2-PAR-4 axis in the tumor microenvironment.Oncotarget. 2017 Jan 3;8(1):1290-1303. doi: 10.18632/oncotarget.13609. Oncotarget. 2017. PMID: 27901488 Free PMC article.
-
FHIT down-regulation was inversely linked to aggressive behaviors and adverse prognosis of gastric cancer: a meta- and bioinformatics analysis.Oncotarget. 2017 Nov 3;8(64):108261-108273. doi: 10.18632/oncotarget.22369. eCollection 2017 Dec 8. Oncotarget. 2017. PMID: 29296239 Free PMC article.
-
Tanshinones induce tumor cell apoptosis via directly targeting FHIT.Sci Rep. 2021 Jun 9;11(1):12217. doi: 10.1038/s41598-021-91708-z. Sci Rep. 2021. PMID: 34108553 Free PMC article.
References
-
- Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rüdiger T, McCue PA, Croce CM, Huebner K. The tumor spectrum in FHIT-deficient mice. Proc Natl Acad Sci USA 2001; 98:10250-5; PMID:11517343; http://dx.doi.org/10.1073/pnas.191345898 - DOI - PMC - PubMed
-
- Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. . The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996; 84:587-97; PMID:8598045; http://dx.doi.org/10.1016/S0092-8674(00)81034-X - DOI - PubMed
-
- Dumon KR, Ishii H, Fong LY, Zanesi N, Fidanza V, Mancini R, Vecchione A, Baffa R, Trapasso F, During MJ, et al. . FHIT gene therapy prevents tumor development in Fhit-deficient mice. Proc Natl Acad Sci USA 2001; 98:3346-51; PMID:11248081; http://dx.doi.org/10.1073/pnas.061020098 - DOI - PMC - PubMed
-
- Trapasso F, Krakowiak A, Cesari R, Arkles J, Yendamuri S, Ishii H, Vecchione A, Kuroki T, Bieganowski P, Pace HC, et al. . Designed FHIT alleles establish that Fhit-induced apoptosis in cancer cells is limited by substrate binding. Proc Natl Acad Sci USA 2003; 100:1592-7; PMID:12574506; http://dx.doi.org/10.1073/pnas.0437915100 - DOI - PMC - PubMed
-
- Sozzi G, Tornielli S, Tagliabue E, Sard L, Pezzella F, Pastorino U, Minoletti F, Pilotti S, Ratcliffe C, Veronese ML, et al. . Absence of Fhit protein in primary lung tumors and cell lines with FHIT gene abnormalities. Cancer Res 1997; 57(23):5207-12; PMID:9393735.12649173 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
