Universal strategies for the DNA-encoding of libraries of small molecules using the chemical ligation of oligonucleotide tags
- PMID: 25483841
- PMCID: PMC4014522
- DOI: 10.4161/adna.27896
Universal strategies for the DNA-encoding of libraries of small molecules using the chemical ligation of oligonucleotide tags
Abstract
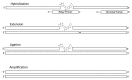
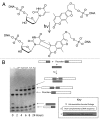
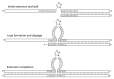
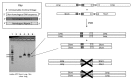
The affinity-mediated selection of large libraries of DNA-encoded small molecules is increasingly being used to initiate drug discovery programs. We present universal methods for the encoding of such libraries using the chemical ligation of oligonucleotides. These methods may be used to record the chemical history of individual library members during combinatorial synthesis processes. We demonstrate three different chemical ligation methods as examples of information recording processes (writing) for such libraries and two different cDNA-generation methods as examples of information retrieval processes (reading) from such libraries. The example writing methods include uncatalyzed and Cu(I)-catalyzed alkyne-azide cycloadditions and a novel photochemical thymidine-psoralen cycloaddition. The first reading method "relay primer-dependent bypass" utilizes a relay primer that hybridizes across a chemical ligation junction embedded in a fixed-sequence and is extended at its 3'-terminus prior to ligation to adjacent oligonucleotides. The second reading method "repeat-dependent bypass" utilizes chemical ligation junctions that are flanked by repeated sequences. The upstream repeat is copied prior to a rearrangement event during which the 3'-terminus of the cDNA hybridizes to the downstream repeat and polymerization continues. In principle these reading methods may be used with any ligation chemistry and offer universal strategies for the encoding (writing) and interpretation (reading) of DNA-encoded chemical libraries.
Keywords: chemical ligation; click chemistry; combinatorial chemistry; drug discovery; in vitro selection; modified nucleotide; photochemistry; polymerase; template-dependent polymerization; unimolecular information recording and retrieval.
Figures




Similar articles
-
Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library.Sci Rep. 2015 Jun 10;5:10916. doi: 10.1038/srep10916. Sci Rep. 2015. PMID: 26061191 Free PMC article.
-
Chemical ligation methods for the tagging of DNA-encoded chemical libraries.Curr Opin Chem Biol. 2015 Jun;26:80-8. doi: 10.1016/j.cbpa.2015.02.015. Epub 2015 Mar 8. Curr Opin Chem Biol. 2015. PMID: 25756406 Review.
-
A fluorescence turn-on detection of copper(II) based on the template-dependent click ligation of oligonucleotides.Talanta. 2015 Jan;132:72-6. doi: 10.1016/j.talanta.2014.08.064. Epub 2014 Sep 6. Talanta. 2015. PMID: 25476281
-
High-throughput synthesis of azide libraries suitable for direct "click" chemistry and in situ screening.Org Biomol Chem. 2009 May 7;7(9):1821-8. doi: 10.1039/b902338k. Epub 2009 Mar 11. Org Biomol Chem. 2009. PMID: 19590777
-
The impact of click chemistry in medicinal chemistry.Expert Opin Drug Discov. 2012 Jun;7(6):489-501. doi: 10.1517/17460441.2012.682725. Epub 2012 May 19. Expert Opin Drug Discov. 2012. PMID: 22607210 Review.
Cited by
-
Recent advances in DNA-encoded dynamic libraries.RSC Chem Biol. 2022 Feb 17;3(4):407-419. doi: 10.1039/d2cb00007e. eCollection 2022 Apr 6. RSC Chem Biol. 2022. PMID: 35441147 Free PMC article. Review.
-
Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries.Molecules. 2021 Mar 22;26(6):1790. doi: 10.3390/molecules26061790. Molecules. 2021. PMID: 33810133 Free PMC article.
-
Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library.Sci Rep. 2015 Jun 10;5:10916. doi: 10.1038/srep10916. Sci Rep. 2015. PMID: 26061191 Free PMC article.
-
Design, preparation, and selection of DNA-encoded dynamic libraries.Chem Sci. 2015 Dec 1;6(12):7097-7104. doi: 10.1039/c5sc02467f. Epub 2015 Sep 11. Chem Sci. 2015. PMID: 28757982 Free PMC article.
-
Converting Double-Stranded DNA-Encoded Libraries (DELs) to Single-Stranded Libraries for More Versatile Selections.ACS Omega. 2022 Mar 24;7(13):11491-11500. doi: 10.1021/acsomega.2c01152. eCollection 2022 Apr 5. ACS Omega. 2022. PMID: 35415338 Free PMC article.
References
-
- Kinoshita Y, Nishigaki K. Enzymatic synthesis of code regions for encoded combinatorial chemistry (ECC) Nucleic Acids Symp Ser. 1995;34:201–2. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
