miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13
- PMID: 25476932
- PMCID: PMC4377070
- DOI: 10.1038/ncomms6671
miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13
Abstract
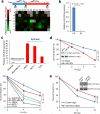
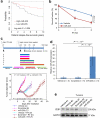
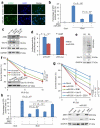
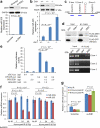
Tumour cells associated with therapy resistance (radioresistance and drug resistance) are likely to give rise to local recurrence and distant metastatic relapse. Recent studies revealed microRNA (miRNA)-mediated regulation of metastasis and epithelial-mesenchymal transition; however, whether specific miRNAs regulate tumour radioresistance and can be exploited as radiosensitizing agents remains unclear. Here we find that miR-205 promotes radiosensitivity and is downregulated in radioresistant subpopulations of breast cancer cells, and that loss of miR-205 is highly associated with poor distant relapse-free survival in breast cancer patients. Notably, therapeutic delivery of miR-205 mimics via nanoliposomes can sensitize the tumour to radiation in a xenograft model. Mechanistically, radiation suppresses miR-205 expression through ataxia telangiectasia mutated (ATM) and zinc finger E-box binding homeobox 1 (ZEB1). Moreover, miR-205 inhibits DNA damage repair by targeting ZEB1 and the ubiquitin-conjugating enzyme Ubc13. These findings identify miR-205 as a radiosensitizing miRNA and reveal a new therapeutic strategy for radioresistant tumours.
Figures
Similar articles
-
ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1.Nat Cell Biol. 2014 Sep;16(9):864-75. doi: 10.1038/ncb3013. Epub 2014 Aug 3. Nat Cell Biol. 2014. PMID: 25086746 Free PMC article.
-
MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer.Int J Cancer. 2014 Sep 15;135(6):1356-68. doi: 10.1002/ijc.28782. Epub 2014 Mar 3. Int J Cancer. 2014. PMID: 24615544
-
Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma.Oncol Rep. 2013 Apr;29(4):1579-87. doi: 10.3892/or.2013.2267. Epub 2013 Jan 31. Oncol Rep. 2013. PMID: 23381389
-
Therapeutic Potential of the miRNA-ATM Axis in the Management of Tumor Radioresistance.Cancer Res. 2020 Jan 15;80(2):139-150. doi: 10.1158/0008-5472.CAN-19-1807. Epub 2019 Nov 25. Cancer Res. 2020. PMID: 31767626 Review.
-
Radio-miRs: a comprehensive view of radioresistance-related microRNAs.Genetics. 2024 Aug 7;227(4):iyae097. doi: 10.1093/genetics/iyae097. Genetics. 2024. PMID: 38963803 Free PMC article. Review.
Cited by
-
MicroRNAs and Their Influence on the ZEB Family: Mechanistic Aspects and Therapeutic Applications in Cancer Therapy.Biomolecules. 2020 Jul 12;10(7):1040. doi: 10.3390/biom10071040. Biomolecules. 2020. PMID: 32664703 Free PMC article. Review.
-
Non-Coding RNAs Associated With Radioresistance in Triple-Negative Breast Cancer.Front Oncol. 2021 Nov 4;11:752270. doi: 10.3389/fonc.2021.752270. eCollection 2021. Front Oncol. 2021. PMID: 34804940 Free PMC article. Review.
-
Decreasing Microtubule Actin Cross-Linking Factor 1 Inhibits Melanoma Metastasis by Decreasing Epithelial to Mesenchymal Transition.Cancer Manag Res. 2020 Jan 29;12:663-673. doi: 10.2147/CMAR.S229156. eCollection 2020. Cancer Manag Res. 2020. PMID: 32099463 Free PMC article.
-
CHFR regulates chemoresistance in triple-negative breast cancer through destabilizing ZEB1.Cell Death Dis. 2021 Aug 30;12(9):820. doi: 10.1038/s41419-021-04114-8. Cell Death Dis. 2021. PMID: 34462429 Free PMC article.
-
Overexpression of Transforming Acidic Coiled Coil‑Containing Protein 3 Reflects Malignant Characteristics and Poor Prognosis of Glioma.Int J Mol Sci. 2017 Mar 4;18(3):235. doi: 10.3390/ijms18030235. Int J Mol Sci. 2017. PMID: 28273854 Free PMC article.
References
-
- Jameel JK, Rao VS, Cawkwell L, Drew PJ. Radioresistance in carcinoma of the breast. Breast. 2004;13:452–460. - PubMed
-
- Horsman MR, et al. Tumor radiosensitizers--current status of development of various approaches: report of an International Atomic Energy Agency meeting. Int J Radiat Oncol Biol Phys. 2006;64:551–561. - PubMed
-
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R43GM100777/GM/NIGMS NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R00 CA138572/CA/NCI NIH HHS/United States
- R01 CA164346/CA/NCI NIH HHS/United States
- P50 CA100632/CA/NCI NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
- R01CA166051/CA/NCI NIH HHS/United States
- R01 CA166051/CA/NCI NIH HHS/United States
- R01CA164346/CA/NCI NIH HHS/United States
- R43 GM100777/GM/NIGMS NIH HHS/United States
- U54 CA151668/CA/NCI NIH HHS/United States
- U54CA151668/CA/NCI NIH HHS/United States
- R00CA138572/CA/NCI NIH HHS/United States
- R01CA181029/CA/NCI NIH HHS/United States
- R44 GM086937/GM/NIGMS NIH HHS/United States
- CA100632/CA/NCI NIH HHS/United States
- R01 CA181029/CA/NCI NIH HHS/United States
- R44GM086937/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

