Shared signatures between rheumatoid arthritis, systemic lupus erythematosus and Sjögren's syndrome uncovered through gene expression meta-analysis
- PMID: 25466291
- PMCID: PMC4295333
- DOI: 10.1186/s13075-014-0489-x
Shared signatures between rheumatoid arthritis, systemic lupus erythematosus and Sjögren's syndrome uncovered through gene expression meta-analysis
Abstract
Introduction: Systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and Sjögren's syndrome (SjS) are inflammatory systemic autoimmune diseases (SADs) that share several clinical and pathological features. The shared biological mechanisms are not yet fully characterized. The objective of this study was to perform a meta-analysis using publicly available gene expression data about the three diseases to identify shared gene expression signatures and overlapping biological processes.
Methods: Previously reported gene expression datasets were selected and downloaded from the Gene Expression Omnibus database. Normalization and initial preprocessing were performed using the statistical programming language R and random effects model-based meta-analysis was carried out using INMEX software. Functional analysis of over- and underexpressed genes was done using the GeneCodis tool.
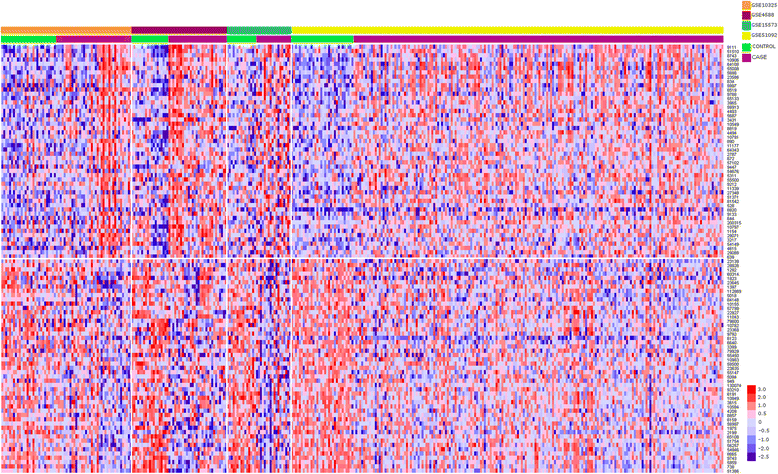
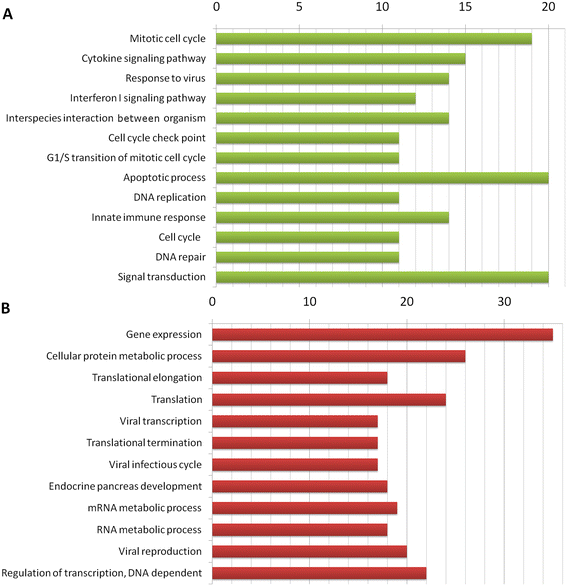
Results: The gene expression meta-analysis revealed a SAD signature composed of 371 differentially expressed genes in patients and healthy controls, 187 of which were underexpressed and 184 overexpressed. Many of these genes have previously been reported as significant biomarkers for individual diseases, but others provide new clues to the shared pathological state. Functional analysis showed that overexpressed genes were involved mainly in immune and inflammatory responses, mitotic cell cycles, cytokine-mediated signaling pathways, apoptotic processes, type I interferon-mediated signaling pathways and responses to viruses. Underexpressed genes were involved primarily in inhibition of protein synthesis.
Conclusions: We define a common gene expression signature for SLE, RA and SjS. The analysis of this signature revealed relevant biological processes that may play important roles in the shared development of these pathologies.
Figures
Similar articles
-
Biosemantics guided gene expression profiling of Sjögren's syndrome: a comparative analysis with systemic lupus erythematosus and rheumatoid arthritis.Arthritis Res Ther. 2017 Aug 17;19(1):192. doi: 10.1186/s13075-017-1400-3. Arthritis Res Ther. 2017. PMID: 28818099 Free PMC article.
-
Germline genetic patterns underlying familial rheumatoid arthritis, systemic lupus erythematosus and primary Sjögren's syndrome highlight T cell-initiated autoimmunity.Ann Rheum Dis. 2020 Feb;79(2):268-275. doi: 10.1136/annrheumdis-2019-215533. Epub 2019 Dec 17. Ann Rheum Dis. 2020. PMID: 31848144
-
Exploring the shared molecular mechanisms between systemic lupus erythematosus and primary Sjögren's syndrome based on integrated bioinformatics and single-cell RNA-seq analysis.Front Immunol. 2023 Aug 8;14:1212330. doi: 10.3389/fimmu.2023.1212330. eCollection 2023. Front Immunol. 2023. PMID: 37614232 Free PMC article.
-
Polyautoimmunity in Sjögren Syndrome.Rheum Dis Clin North Am. 2016 Aug;42(3):457-72. doi: 10.1016/j.rdc.2016.03.005. Epub 2016 Jun 21. Rheum Dis Clin North Am. 2016. PMID: 27431348 Review.
-
[Difficulties in differential diagnosis of Sjögren's syndrome and systemic lupus erythematosus].Przegl Lek. 2006;63(5):278-83. Przegl Lek. 2006. PMID: 17036505 Review. Polish.
Cited by
-
Detection of Genetic Overlap Between Rheumatoid Arthritis and Systemic Lupus Erythematosus Using GWAS Summary Statistics.Front Genet. 2021 Mar 18;12:656545. doi: 10.3389/fgene.2021.656545. eCollection 2021. Front Genet. 2021. PMID: 33815486 Free PMC article.
-
Moving towards a molecular taxonomy of autoimmune rheumatic diseases.Nat Rev Rheumatol. 2018 Jan 24;14(2):75-93. doi: 10.1038/nrrheum.2017.220. Nat Rev Rheumatol. 2018. PMID: 29362467 Review.
-
Transcriptomic characterization of differential gene expression in oral squamous cell carcinoma: a meta-analysis of publicly available microarray data sets.Tumour Biol. 2016 Dec;37:15913–15924. doi: 10.1007/s13277-016-5439-6. Epub 2016 Oct 4. Tumour Biol. 2016. PMID: 27704359
-
Deciphering novel common gene signatures for rheumatoid arthritis and systemic lupus erythematosus by integrative analysis of transcriptomic profiles.PLoS One. 2023 Mar 16;18(3):e0281637. doi: 10.1371/journal.pone.0281637. eCollection 2023. PLoS One. 2023. PMID: 36928613 Free PMC article.
-
Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology.Pharmaceuticals (Basel). 2018 Jun 22;11(3):61. doi: 10.3390/ph11030061. Pharmaceuticals (Basel). 2018. PMID: 29932108 Free PMC article.
References
-
- van ’t Veer LJ, Bernards R: Enabling personalized cancer medicine through analysis of gene-expression patterns.Nature 2008, 452:564–570. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

