Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis
- PMID: 25445340
- PMCID: PMC4465284
- DOI: 10.1016/j.virusres.2014.11.021
Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis
Abstract
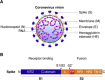
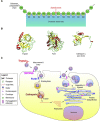
Coronaviruses are a large group of enveloped, single-stranded positive-sense RNA viruses that infect a wide range of avian and mammalian species, including humans. The emergence of deadly human coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) have bolstered research in these viral and often zoonotic pathogens. While coronavirus cell and tissue tropism, host range, and pathogenesis are initially controlled by interactions between the spike envelope glycoprotein and host cell receptor, it is becoming increasingly apparent that proteolytic activation of spike by host cell proteases also plays a critical role. Coronavirus spike proteins are the main determinant of entry as they possess both receptor binding and fusion functions. Whereas binding to the host cell receptor is an essential first step in establishing infection, the proteolytic activation step is often critical for the fusion function of spike, as it allows for controlled release of the fusion peptide into target cellular membranes. Coronaviruses have evolved multiple strategies for proteolytic activation of spike, and a large number of host proteases have been shown to proteolytically process the spike protein. These include, but are not limited to, endosomal cathepsins, cell surface transmembrane protease/serine (TMPRSS) proteases, furin, and trypsin. This review focuses on the diversity of strategies coronaviruses have evolved to proteolytically activate their fusion protein during spike protein biosynthesis and the critical entry step of their life cycle, and highlights important findings on how proteolytic activation of coronavirus spike influences tissue and cell tropism, host range and pathogenicity.
Keywords: Cleavage activation; Coronavirus; Pathogenesis; Protease; Spike protein; Tropism.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Lysosomal Proteases Are a Determinant of Coronavirus Tropism.J Virol. 2018 Nov 27;92(24):e01504-18. doi: 10.1128/JVI.01504-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258004 Free PMC article.
-
Middle East Respiratory Syndrome Coronavirus Spike Protein Is Not Activated Directly by Cellular Furin during Viral Entry into Target Cells.J Virol. 2018 Sep 12;92(19):e00683-18. doi: 10.1128/JVI.00683-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021905 Free PMC article.
-
Genome-wide bioinformatics analysis of human protease capacity for proteolytic cleavage of the SARS-CoV-2 spike glycoprotein.Microbiol Spectr. 2024 Feb 6;12(2):e0353023. doi: 10.1128/spectrum.03530-23. Epub 2024 Jan 8. Microbiol Spectr. 2024. PMID: 38189333 Free PMC article.
-
Coronavirus Spike Protein and Tropism Changes.Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review.
-
Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond.Trends Microbiol. 2015 Aug;23(8):468-78. doi: 10.1016/j.tim.2015.06.003. Epub 2015 Jul 21. Trends Microbiol. 2015. PMID: 26206723 Free PMC article. Review.
Cited by
-
Structural analysis of experimental drugs binding to the SARS-CoV-2 target TMPRSS2.J Mol Graph Model. 2020 Nov;100:107710. doi: 10.1016/j.jmgm.2020.107710. Epub 2020 Aug 11. J Mol Graph Model. 2020. PMID: 32829149 Free PMC article.
-
Inhibitors of SARS-CoV-2 Entry: Current and Future Opportunities.J Med Chem. 2020 Nov 12;63(21):12256-12274. doi: 10.1021/acs.jmedchem.0c00502. Epub 2020 Jun 25. J Med Chem. 2020. PMID: 32539378 Free PMC article. Review.
-
Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor.Biosensors (Basel). 2022 Oct 17;12(10):888. doi: 10.3390/bios12100888. Biosensors (Basel). 2022. PMID: 36291025 Free PMC article.
-
COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells.Genes (Basel). 2020 Aug 27;11(9):1010. doi: 10.3390/genes11091010. Genes (Basel). 2020. PMID: 32867305 Free PMC article.
-
Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19.iScience. 2021 Mar 19;24(3):102254. doi: 10.1016/j.isci.2021.102254. Epub 2021 Mar 1. iScience. 2021. PMID: 33681723 Free PMC article.
References
-
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., de Wit E., Munster V.J., Hensley L.E., Zalmout I.S., Kapoor A., Epstein J.H., Karesh W.B., Daszak P., Mohammed O.B., Lipkin W.I. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5(2):e00884-14. - PMC - PubMed
-
- Backes B.J., Harris J.L., Leonetti F., Craik C.S., Ellman J.A. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 2000;18(2):187–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

