The centromere: epigenetic control of chromosome segregation during mitosis
- PMID: 25414369
- PMCID: PMC4292168
- DOI: 10.1101/cshperspect.a015818
The centromere: epigenetic control of chromosome segregation during mitosis
Abstract
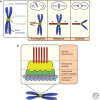
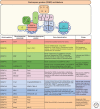
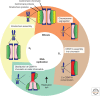
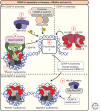
A fundamental challenge for the survival of all organisms is maintaining the integrity of the genome in all cells. Cells must therefore segregate their replicated genome equally during each cell division. Eukaryotic organisms package their genome into a number of physically distinct chromosomes, which replicate during S phase and condense during prophase of mitosis to form paired sister chromatids. During mitosis, cells form a physical connection between each sister chromatid and microtubules of the mitotic spindle, which segregate one copy of each chromatid to each new daughter cell. The centromere is the DNA locus on each chromosome that creates the site of this connection. In this review, we present a brief history of centromere research and discuss our current knowledge of centromere establishment, maintenance, composition, structure, and function in mitosis.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

Similar articles
-
Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation.J Cell Biol. 2011 Sep 5;194(5):721-35. doi: 10.1083/jcb.201010118. Epub 2011 Aug 29. J Cell Biol. 2011. PMID: 21875947 Free PMC article.
-
Centromere Dysfunction Compromises Mitotic Spindle Pole Integrity.Curr Biol. 2019 Sep 23;29(18):3072-3080.e5. doi: 10.1016/j.cub.2019.07.052. Epub 2019 Sep 5. Curr Biol. 2019. PMID: 31495582
-
Condensin in Chromatid Cohesion and Segregation.Cytogenet Genome Res. 2015;147(4):212-6. doi: 10.1159/000444868. Epub 2016 Mar 22. Cytogenet Genome Res. 2015. PMID: 26998746 Review.
-
Epigenetic regulation of centromere formation and kinetochore function.Biochem Cell Biol. 2006 Aug;84(4):605-18. doi: 10.1139/o06-080. Biochem Cell Biol. 2006. PMID: 16936832 Review.
-
Kinetochore assembly and function through the cell cycle.Chromosoma. 2016 Sep;125(4):645-59. doi: 10.1007/s00412-016-0608-3. Epub 2016 Jul 4. Chromosoma. 2016. PMID: 27376724 Review.
Cited by
-
Centromeres under Pressure: Evolutionary Innovation in Conflict with Conserved Function.Genes (Basel). 2020 Aug 10;11(8):912. doi: 10.3390/genes11080912. Genes (Basel). 2020. PMID: 32784998 Free PMC article. Review.
-
CDK phosphorylation of Xenopus laevis M18BP1 promotes its metaphase centromere localization.EMBO J. 2019 Feb 15;38(4):e100093. doi: 10.15252/embj.2018100093. Epub 2019 Jan 2. EMBO J. 2019. PMID: 30606714 Free PMC article.
-
The cell cycle, cancer development and therapy.Mol Biol Rep. 2022 Nov;49(11):10875-10883. doi: 10.1007/s11033-022-07788-1. Epub 2022 Aug 5. Mol Biol Rep. 2022. PMID: 35931874 Review.
-
Sequence, Chromatin and Evolution of Satellite DNA.Int J Mol Sci. 2021 Apr 21;22(9):4309. doi: 10.3390/ijms22094309. Int J Mol Sci. 2021. PMID: 33919233 Free PMC article. Review.
-
CENP-N promotes the compaction of centromeric chromatin.Nat Struct Mol Biol. 2022 Apr;29(4):403-413. doi: 10.1038/s41594-022-00758-y. Epub 2022 Apr 14. Nat Struct Mol Biol. 2022. PMID: 35422519 Free PMC article.
References
-
- Bade D, Pauleau AL, Wendler A, Erhardt S 2014. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell 28: 508–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
