Transmembrane domains of attraction on the TSH receptor
- PMID: 25406938
- PMCID: PMC4298320
- DOI: 10.1210/en.2014-1509
Transmembrane domains of attraction on the TSH receptor
Abstract
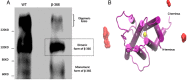
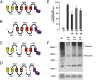
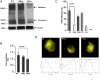
The TSH receptor (TSHR) has the propensity to form dimers and oligomers. Our data using ectodomain-truncated TSHRs indicated that the predominant interfaces for oligomerization reside in the transmembrane (TM) domain. To map the potentially interacting residues, we first performed in silico studies of the TSHR transmembrane domain using a homology model and using Brownian dynamics (BD). The cluster of dimer conformations obtained from BD analysis indicated that TM1 made contact with TM4 and two residues in TM2 made contact with TM5. To confirm the proximity of these contact residues, we then generated cysteine mutants at all six contact residues predicted by the BD analysis and performed cysteine cross-linking studies. These results showed that the predicted helices in the protomer were indeed involved in proximity interactions. Furthermore, an alternative experimental approach, receptor truncation experiments and LH receptor sequence substitution experiments, identified TM1 harboring a major region involved in TSHR oligomerization, in agreement with the conclusion from the cross-linking studies. Point mutations of the predicted interacting residues did not yield a substantial decrease in oligomerization, unlike the truncation of the TM1, so we concluded that constitutive oligomerization must involve interfaces forming domains of attraction in a cooperative manner that is not dominated by interactions between specific residues.
Figures
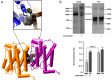
Similar articles
-
Effects of mutations involving cysteine residues distal to the S281HCC motif at the C-terminus on the functional characteristics of a truncated ectodomain-only thyrotropin receptor anchored on glycosylphosphatidyl-inositol.Thyroid. 2008 Dec;18(12):1313-9. doi: 10.1089/thy.2008.0240. Thyroid. 2008. PMID: 18976165
-
A tyrosine residue on the TSH receptor stabilizes multimer formation.PLoS One. 2010 Feb 26;5(2):e9449. doi: 10.1371/journal.pone.0009449. PLoS One. 2010. PMID: 20195479 Free PMC article.
-
Molecular interactions between the TSH receptor and a Thyroid-stimulating monoclonal autoantibody.Thyroid. 2007 Aug;17(8):699-706. doi: 10.1089/thy.2007.0041. Thyroid. 2007. PMID: 17725428
-
Evidence that the C terminus of the A subunit suppresses thyrotropin receptor constitutive activity.Endocrinology. 2003 Sep;144(9):3821-7. doi: 10.1210/en.2003-0430. Endocrinology. 2003. PMID: 12933653
-
Minireview: structural and functional evolution of the thyrotropin receptor.Endocrinology. 2004 Sep;145(9):4048-57. doi: 10.1210/en.2004-0437. Epub 2004 Jul 1. Endocrinology. 2004. PMID: 15231707 Review.
Cited by
-
A Modifying Autoantigen in Graves' Disease.Endocrinology. 2019 May 1;160(5):1008-1020. doi: 10.1210/en.2018-01048. Endocrinology. 2019. PMID: 30822352 Free PMC article.
-
Structure and activation of the TSH receptor transmembrane domain.Auto Immun Highlights. 2017 Dec;8(1):2. doi: 10.1007/s13317-016-0090-1. Epub 2016 Dec 5. Auto Immun Highlights. 2017. PMID: 27921237 Free PMC article.
-
Structural-Functional Features of the Thyrotropin Receptor: A Class A G-Protein-Coupled Receptor at Work.Front Endocrinol (Lausanne). 2017 Apr 24;8:86. doi: 10.3389/fendo.2017.00086. eCollection 2017. Front Endocrinol (Lausanne). 2017. PMID: 28484426 Free PMC article. Review.
-
Targeting the thyroid-stimulating hormone receptor with small molecule ligands and antibodies.Expert Opin Ther Targets. 2015 Jun;19(6):835-47. doi: 10.1517/14728222.2015.1018181. Epub 2015 Mar 13. Expert Opin Ther Targets. 2015. PMID: 25768836 Free PMC article. Review.
-
Structure of full-length TSH receptor in complex with antibody K1-70™.J Mol Endocrinol. 2022 Dec 7;70(1):e220120. doi: 10.1530/JME-22-0120. Print 2023 Jan 1. J Mol Endocrinol. 2022. PMID: 36069797 Free PMC article.
References
-
- Ciruela F, Vallano A, Arnau JM, et al. G protein-coupled receptor oligomerization for what? J Recept Signal Transduct Res. 2010;30(5):322–330. - PubMed
-
- Javitch JA. The ants go marching two by two: oligomeric structure of G-protein-coupled receptors. Mol Pharmacol. 2004;66(5):1077–1082. - PubMed
-
- Lee C, Ji I, Ryu K, Song Y, Conn PM, Ji TH. Two defective heterozygous luteinizing hormone receptors can rescue hormone action. J Biol Chem. 2002;277(18):15795–15800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

