Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids
- PMID: 25402640
- PMCID: PMC4264194
- DOI: 10.3390/ijms151120753
Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids
Abstract
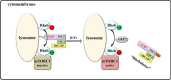
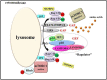
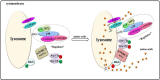
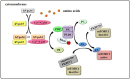
Mammalian target of rapamycin (mTOR), which is now referred to as mechanistic target of rapamycin, integrates many signals, including those from growth factors, energy status, stress, and amino acids, to regulate cell growth and proliferation, protein synthesis, protein degradation, and other physiological and biochemical processes. The mTOR-Rheb-TSC-TBC complex co-localizes to the lysosome and the phosphorylation of TSC-TBC effects the dissociation of the complex from the lysosome and activates Rheb. GTP-bound Rheb potentiates the catalytic activity of mTORC1. Under conditions with growth factors and amino acids, v-ATPase, Ragulator, Rag GTPase, Rheb, hVps34, PLD1, and PA have important but disparate effects on mTORC1 activation. In this review, we introduce five models of mTORC1 activation by growth factors and amino acids to provide a comprehensive theoretical foundation for future research.
Figures
Similar articles
-
Rheb and Rags come together at the lysosome to activate mTORC1.Biochem Soc Trans. 2013 Aug;41(4):951-5. doi: 10.1042/BST20130037. Biochem Soc Trans. 2013. PMID: 23863162 Review.
-
Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity.Elife. 2016 Jan 7;5:e11058. doi: 10.7554/eLife.11058. Elife. 2016. PMID: 26742086 Free PMC article.
-
Requirement for lysosomal localization of mTOR for its activation differs between leucine and other amino acids.Cell Signal. 2014 Sep;26(9):1918-27. doi: 10.1016/j.cellsig.2014.04.019. Epub 2014 May 2. Cell Signal. 2014. PMID: 24793303
-
Key mediators of intracellular amino acids signaling to mTORC1 activation.Amino Acids. 2015 May;47(5):857-67. doi: 10.1007/s00726-015-1937-x. Epub 2015 Feb 21. Amino Acids. 2015. PMID: 25701492 Review.
-
Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging.J Biol Chem. 2014 Jan 31;289(5):2658-74. doi: 10.1074/jbc.M113.528505. Epub 2013 Dec 11. J Biol Chem. 2014. PMID: 24337580 Free PMC article.
Cited by
-
It's all about talking: two-way communication between proteasomal and lysosomal degradation pathways via ubiquitin.Am J Physiol Cell Physiol. 2016 Aug 1;311(2):C166-78. doi: 10.1152/ajpcell.00074.2016. Epub 2016 May 25. Am J Physiol Cell Physiol. 2016. PMID: 27225656 Free PMC article.
-
Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma.Leukemia. 2017 Jan;31(1):83-91. doi: 10.1038/leu.2016.175. Epub 2016 Jun 16. Leukemia. 2017. PMID: 27389057 Free PMC article.
-
Chromosome-associated protein D3 promotes bacterial clearance in human intestinal epithelial cells by repressing expression of amino acid transporters.Gastroenterology. 2015 Jun;148(7):1405-1416.e3. doi: 10.1053/j.gastro.2015.02.013. Epub 2015 Feb 18. Gastroenterology. 2015. PMID: 25701737 Free PMC article.
-
TNFAIP8L2/TIPE2 impairs autolysosome reformation via modulating the RAC1-MTORC1 axis.Autophagy. 2021 Jun;17(6):1410-1425. doi: 10.1080/15548627.2020.1761748. Epub 2020 May 28. Autophagy. 2021. PMID: 32460619 Free PMC article.
-
The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases.Front Mol Neurosci. 2019 May 21;12:121. doi: 10.3389/fnmol.2019.00121. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31213978 Free PMC article. Review.
References
-
- Wullschleger S., Loewith R., Hall M.N. Tor signaling in growth and metabolism. Cell. 2006;124:471–484. - PubMed
-
- Brown E.J., Albers M.W., Shin T.B., Ichikawa K., Keith C.T., Lane W.S., Schreiber S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. - PubMed
-
- Heitman J., Movva N.R., Hall M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

