Three-dimensional structure of a protozoal double-stranded RNA virus that infects the enteric pathogen Giardia lamblia
- PMID: 25378500
- PMCID: PMC4300635
- DOI: 10.1128/JVI.02745-14
Three-dimensional structure of a protozoal double-stranded RNA virus that infects the enteric pathogen Giardia lamblia
Abstract
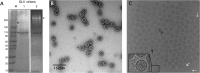
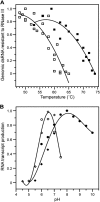
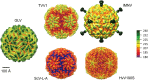
Giardia lamblia virus (GLV) is a small, nonenveloped, nonsegmented double-stranded RNA (dsRNA) virus infecting Giardia lamblia, the most common protozoan pathogen of the human intestine and a major agent of waterborne diarrheal disease worldwide. GLV (genus Giardiavirus) is a member of family Totiviridae, along with several other groups of protozoal or fungal viruses, including Leishmania RNA viruses and Trichomonas vaginalis viruses. Interestingly, GLV is more closely related than other Totiviridae members to a group of recently discovered metazoan viruses that includes penaeid shrimp infectious myonecrosis virus (IMNV). Moreover, GLV is the only known protozoal dsRNA virus that can transmit efficiently by extracellular means, also like IMNV. In this study, we used transmission electron cryomicroscopy and icosahedral image reconstruction to examine the GLV virion at an estimated resolution of 6.0 Å. Its outermost diameter is 485 Å, making it the largest totivirus capsid analyzed to date. Structural comparisons of GLV and other totiviruses highlighted a related "T=2" capsid organization and a conserved helix-rich fold in the capsid subunits. In agreement with its unique capacity as a protozoal dsRNA virus to survive and transmit through extracellular environments, GLV was found to be more thermoresistant than Trichomonas vaginalis virus 1, but no specific protein machinery to mediate cell entry, such as the fiber complexes in IMNV, could be localized. These and other structural and biochemical findings provide a basis for future work to dissect the cell entry mechanism of GLV into a "primitive" (early-branching) eukaryotic host and an important enteric pathogen of humans.
Importance: Numerous pathogenic bacteria, including Corynebacterium diphtheriae, Salmonella enterica, and Vibrio cholerae, are infected with lysogenic bacteriophages that contribute significantly to bacterial virulence. In line with this phenomenon, several pathogenic protozoa, including Giardia lamblia, Leishmania species, and Trichomonas vaginalis are persistently infected with dsRNA viruses, and growing evidence indicates that at least some of these protozoal viruses can likewise enhance the pathogenicity of their hosts. Understanding of these protozoal viruses, however, lags far behind that of many bacteriophages. Here, we investigated the dsRNA virus that infects the widespread enteric parasite Giardia lamblia. Using electron cryomicroscopy and icosahedral image reconstruction, we determined the virion structure of Giardia lamblia virus, obtaining new information relating to its assembly, stability, functions in cell entry and transcription, and similarities and differences with other dsRNA viruses. The results of our study set the stage for further mechanistic work on the roles of these viruses in protozoal virulence.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.PLoS Pathog. 2024 Apr 10;20(4):e1012140. doi: 10.1371/journal.ppat.1012140. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38598600 Free PMC article.
-
Identification of virus-specific vesicles in Giardiavirus-infected Giardia lamblia.J Microbiol Immunol Infect. 2000 Mar;33(1):9-13. J Microbiol Immunol Infect. 2000. PMID: 10806957
-
Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.mBio. 2021 Mar 30;12(2):e02924-20. doi: 10.1128/mBio.02924-20. mBio. 2021. PMID: 33785622 Free PMC article.
-
Giardiavirus: an update.Parasitol Res. 2021 Jun;120(6):1943-1948. doi: 10.1007/s00436-021-07167-y. Epub 2021 May 6. Parasitol Res. 2021. PMID: 33956215 Review.
-
Recent advances in molecular biology of parasitic viruses.Infect Disord Drug Targets. 2014;14(3):155-67. doi: 10.2174/1871526514666140713160905. Infect Disord Drug Targets. 2014. PMID: 25019235 Review.
Cited by
-
Microbial Matryoshka: Addressing the Relationship between Pathogenic Flagellated Protozoans and Their RNA Viral Endosymbionts (Family Totiviridae).Vet Sci. 2024 Jul 17;11(7):321. doi: 10.3390/vetsci11070321. Vet Sci. 2024. PMID: 39058005 Free PMC article. Review.
-
High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.PLoS Pathog. 2024 Apr 10;20(4):e1012140. doi: 10.1371/journal.ppat.1012140. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38598600 Free PMC article.
-
Metagenomic of Liver Tissue Identified at Least Two Genera of Totivirus-like Viruses in Molossus molossus Bats.Microorganisms. 2024 Jan 19;12(1):206. doi: 10.3390/microorganisms12010206. Microorganisms. 2024. PMID: 38276191 Free PMC article.
-
Multiple Regulations of Parasitic Protozoan Viruses: A Double-Edged Sword for Protozoa.mBio. 2023 Feb 28;14(1):e0264222. doi: 10.1128/mbio.02642-22. Epub 2023 Jan 12. mBio. 2023. PMID: 36633419 Free PMC article. Review.
-
Characterisation of Trichomonas vaginalis Isolates Collected from Patients in Vienna between 2019 and 2021.Int J Mol Sci. 2022 Oct 17;23(20):12422. doi: 10.3390/ijms232012422. Int J Mol Sci. 2022. PMID: 36293276 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

