A conserved regulatory module at the C terminus of the papillomavirus E1 helicase domain controls E1 helicase assembly
- PMID: 25378487
- PMCID: PMC4300631
- DOI: 10.1128/JVI.01903-14
A conserved regulatory module at the C terminus of the papillomavirus E1 helicase domain controls E1 helicase assembly
Abstract
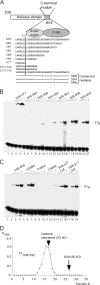
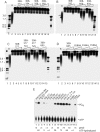
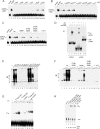
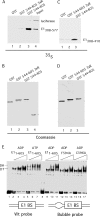
Viruses frequently combine multiple activities into one polypeptide to conserve coding capacity. This strategy creates regulatory challenges to ascertain that the combined activities are compatible and do not interfere with each other. The papillomavirus E1 protein, as many other helicases, has the intrinsic ability to form hexamers and double hexamers (DH) that serve as the replicative DNA helicase. However, E1 also has the more unusual ability to generate local melting by forming a double trimer (DT) complex that can untwist the double-stranded origin of DNA replication (ori) DNA in preparation for DH formation. Here we describe a switching mechanism that allows the papillomavirus E1 protein to form these two different kinds of oligomers and to transition between them. We show that a conserved regulatory module attached to the E1 helicase domain blocks hexamer and DH formation and promotes DT formation. In the presence of the appropriate trigger, the inhibitory effect of the regulatory module is relieved and the transition to DH formation can occur.
Importance: This study provides a mechanistic understanding into how a multifunctional viral polypeptide can provide different, seemingly incompatible activities. A conserved regulatory sequence module attached to the AAA+ helicase domain in the papillomavirus E1 protein allows the formation of different oligomers with different biochemical activities.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Assembly of a double hexameric helicase.Mol Cell. 2005 Nov 11;20(3):377-89. doi: 10.1016/j.molcel.2005.09.020. Mol Cell. 2005. PMID: 16285920
-
Adjacent residues in the E1 initiator beta-hairpin define different roles of the beta-hairpin in Ori melting, helicase loading, and helicase activity.Mol Cell. 2007 Mar 23;25(6):825-37. doi: 10.1016/j.molcel.2007.02.009. Mol Cell. 2007. PMID: 17386260
-
Mechanistic analysis of local ori melting and helicase assembly by the papillomavirus E1 protein.Mol Cell. 2011 Sep 2;43(5):776-87. doi: 10.1016/j.molcel.2011.06.026. Mol Cell. 2011. PMID: 21884978 Free PMC article.
-
Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein.J Virol. 2003 May;77(9):5178-91. doi: 10.1128/jvi.77.9.5178-5191.2003. J Virol. 2003. PMID: 12692220 Free PMC article.
-
The E1 proteins.Virology. 2013 Oct;445(1-2):35-56. doi: 10.1016/j.virol.2013.07.020. Epub 2013 Sep 10. Virology. 2013. PMID: 24029589 Free PMC article. Review.
Cited by
-
Human Papillomavirus Replication Regulation by Acetylation of a Conserved Lysine in the E2 Protein.J Virol. 2018 Jan 17;92(3):e01912-17. doi: 10.1128/JVI.01912-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142126 Free PMC article.
-
Active DNA unwinding and transport by a membrane-adapted helicase nanopore.Nat Commun. 2019 Nov 8;10(1):5083. doi: 10.1038/s41467-019-13047-y. Nat Commun. 2019. PMID: 31704937 Free PMC article.
-
Requirement for the E1 Helicase C-Terminal Domain in Papillomavirus DNA Replication In Vivo.J Virol. 2016 Jan 6;90(6):3198-211. doi: 10.1128/JVI.03127-15. J Virol. 2016. PMID: 26739052 Free PMC article.
-
Mechanisms of hexameric helicases.Crit Rev Biochem Mol Biol. 2021 Dec;56(6):621-639. doi: 10.1080/10409238.2021.1954597. Epub 2021 Aug 17. Crit Rev Biochem Mol Biol. 2021. PMID: 34404299 Free PMC article. Review.
-
Papillomaviruses: a systematic review.Genet Mol Biol. 2017 Jan-Mar;40(1):1-21. doi: 10.1590/1678-4685-GMB-2016-0128. Epub 2017 Feb 16. Genet Mol Biol. 2017. PMID: 28212457 Free PMC article.
References
-
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9:27–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

