Genetic and epigenetic fine mapping of causal autoimmune disease variants
- PMID: 25363779
- PMCID: PMC4336207
- DOI: 10.1038/nature13835
Genetic and epigenetic fine mapping of causal autoimmune disease variants
Abstract
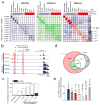
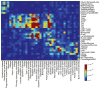
Genome-wide association studies have identified loci underlying human diseases, but the causal nucleotide changes and mechanisms remain largely unknown. Here we developed a fine-mapping algorithm to identify candidate causal variants for 21 autoimmune diseases from genotyping data. We integrated these predictions with transcription and cis-regulatory element annotations, derived by mapping RNA and chromatin in primary immune cells, including resting and stimulated CD4(+) T-cell subsets, regulatory T cells, CD8(+) T cells, B cells, and monocytes. We find that ∼90% of causal variants are non-coding, with ∼60% mapping to immune-cell enhancers, many of which gain histone acetylation and transcribe enhancer-associated RNA upon immune stimulation. Causal variants tend to occur near binding sites for master regulators of immune differentiation and stimulus-dependent gene activation, but only 10-20% directly alter recognizable transcription factor binding motifs. Rather, most non-coding risk variants, including those that alter gene expression, affect non-canonical sequence determinants not well-explained by current gene regulatory models.
Conflict of interest statement
Figures


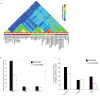
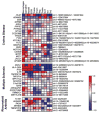
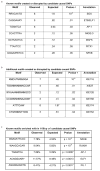
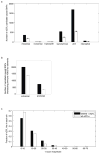
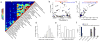



Comment in
-
Genetics: Mapping autoimmune disease epigenetics: what's on the horizon?Nat Rev Rheumatol. 2015 Mar;11(3):131-2. doi: 10.1038/nrrheum.2014.210. Epub 2014 Dec 16. Nat Rev Rheumatol. 2015. PMID: 25512011 No abstract available.
-
Take your PICS: moving from GWAS to immune function.Immunity. 2014 Dec 18;41(6):883-5. doi: 10.1016/j.immuni.2014.12.014. Immunity. 2014. PMID: 25526303
-
Causal variants in autoimmune disease: a commentary on a recent published fine-mapping algorithm analysis in genome-wide association studies study.Ann Transl Med. 2017 Mar;5(6):151. doi: 10.21037/atm.2017.02.26. Ann Transl Med. 2017. PMID: 28462231 Free PMC article. No abstract available.
Similar articles
-
Chronic lymphocytic leukemia (CLL) risk is mediated by multiple enhancer variants within CLL risk loci.Hum Mol Genet. 2020 Sep 29;29(16):2761-2774. doi: 10.1093/hmg/ddaa165. Hum Mol Genet. 2020. PMID: 32744316 Free PMC article.
-
Making the most of new genetic risk factors - genetic and epigenetic fine mapping of causal autoimmune disease variants.Clin Res Hepatol Gastroenterol. 2015 Sep;39(4):408-11. doi: 10.1016/j.clinre.2015.05.002. Epub 2015 Jul 6. Clin Res Hepatol Gastroenterol. 2015. PMID: 26160476 No abstract available.
-
Joint Bayesian inference of risk variants and tissue-specific epigenomic enrichments across multiple complex human diseases.Nucleic Acids Res. 2016 Oct 14;44(18):e144. doi: 10.1093/nar/gkw627. Epub 2016 Jul 12. Nucleic Acids Res. 2016. PMID: 27407109 Free PMC article.
-
The complexity of epigenetic diseases.J Pathol. 2016 Jan;238(2):333-44. doi: 10.1002/path.4647. Epub 2015 Nov 17. J Pathol. 2016. PMID: 26419725 Free PMC article. Review.
-
Fine mapping with epigenetic information and 3D structure.Semin Immunopathol. 2022 Jan;44(1):115-125. doi: 10.1007/s00281-021-00906-4. Epub 2022 Jan 12. Semin Immunopathol. 2022. PMID: 35022890 Free PMC article. Review.
Cited by
-
eQTL in diseased colon tissue identifies novel target genes associated with IBD.bioRxiv [Preprint]. 2024 Oct 17:2024.10.14.618229. doi: 10.1101/2024.10.14.618229. bioRxiv. 2024. PMID: 39464142 Free PMC article. Preprint.
-
Artificial Intelligence in Epigenetic Studies: Shedding Light on Rare Diseases.Front Mol Biosci. 2021 May 5;8:648012. doi: 10.3389/fmolb.2021.648012. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026829 Free PMC article. Review.
-
CD4:CD8 lymphocyte ratio as a quantitative measure of immunologic health in HIV-1 infection: findings from an African cohort with prospective data.Front Microbiol. 2015 Jul 1;6:670. doi: 10.3389/fmicb.2015.00670. eCollection 2015. Front Microbiol. 2015. PMID: 26191056 Free PMC article.
-
Review of multi-omics data resources and integrative analysis for human brain disorders.Brief Funct Genomics. 2021 Jul 17;20(4):223-234. doi: 10.1093/bfgp/elab024. Brief Funct Genomics. 2021. PMID: 33969380 Free PMC article. Review.
-
Cell-Based Therapy and Genome Editing as Emerging Therapeutic Approaches to Treat Rheumatoid Arthritis.Cells. 2024 Jul 30;13(15):1282. doi: 10.3390/cells13151282. Cells. 2024. PMID: 39120313 Free PMC article. Review.
References
-
- Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007019/AI/NIAID NIH HHS/United States
- U19 AI070352/AI/NIAID NIH HHS/United States
- AI070352/AI/NIAID NIH HHS/United States
- NS24247/NS/NINDS NIH HHS/United States
- R37 NS024247/NS/NINDS NIH HHS/United States
- R01 NS067305/NS/NINDS NIH HHS/United States
- R01 NS024247/NS/NINDS NIH HHS/United States
- NS067305/NS/NINDS NIH HHS/United States
- ES017155/ES/NIEHS NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- HG004570/HG/NHGRI NIH HHS/United States
- P01 AI045757/AI/NIAID NIH HHS/United States
- AI046130/AI/NIAID NIH HHS/United States
- U19 AI046130/AI/NIAID NIH HHS/United States
- U01 ES017155/ES/NIEHS NIH HHS/United States
- GM093080/GM/NIGMS NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AI039671/AI/NIAID NIH HHS/United States
- U54 HG006991/HG/NHGRI NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- AI045757/AI/NIAID NIH HHS/United States
- U54 HG004570/HG/NHGRI NIH HHS/United States
- RC2 GM093080/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

