The cullin-4 complex DCDC does not require E3 ubiquitin ligase elements to control heterochromatin in Neurospora crassa
- PMID: 25362134
- PMCID: PMC4279019
- DOI: 10.1128/EC.00212-14
The cullin-4 complex DCDC does not require E3 ubiquitin ligase elements to control heterochromatin in Neurospora crassa
Abstract
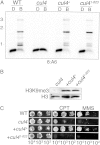
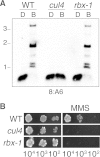
The cullin-4 (CUL4) complex DCDC (DIM-5/-7/-9/CUL4/DDB1 complex) is essential for DNA methylation and heterochromatin formation in Neurospora crassa. Cullins form the scaffold of cullin-RING E3 ubiquitin ligases (CRLs) and are modified by the covalent attachment of NEDD8, a ubiquitin-like protein that regulates the stability and activity of CRLs. We report that neddylation is not required for CUL4-dependent DNA methylation or heterochromatin formation but is required for the DNA repair functions. Moreover, the RING domain protein RBX1 and a segment of the CUL4 C terminus that normally interacts with RBX1, the E2 ligase, CAND1, and CSN are dispensable for DNA methylation and heterochromatin formation by DCDC. Our study provides evidence for the noncanonical functions of core CRL components.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa.J Biol Chem. 2010 Feb 12;285(7):4355-65. doi: 10.1074/jbc.M109.034710. Epub 2009 Nov 30. J Biol Chem. 2010. PMID: 19948733 Free PMC article.
-
DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa.PLoS Genet. 2010 Sep 23;6(9):e1001132. doi: 10.1371/journal.pgen.1001132. PLoS Genet. 2010. PMID: 20885793 Free PMC article.
-
DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC.PLoS Genet. 2010 Nov 4;6(11):e1001196. doi: 10.1371/journal.pgen.1001196. PLoS Genet. 2010. PMID: 21079689 Free PMC article.
-
Protection of cullin-RING E3 ligases by CSN-UBP12.Trends Cell Biol. 2006 Jul;16(7):362-9. doi: 10.1016/j.tcb.2006.05.001. Epub 2006 Jun 9. Trends Cell Biol. 2006. PMID: 16762551 Review.
-
Structural regulation of cullin-RING ubiquitin ligase complexes.Curr Opin Struct Biol. 2011 Apr;21(2):257-64. doi: 10.1016/j.sbi.2011.01.003. Epub 2011 Feb 1. Curr Opin Struct Biol. 2011. PMID: 21288713 Free PMC article. Review.
Cited by
-
Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9598-E9607. doi: 10.1073/pnas.1715049114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078403 Free PMC article.
-
Genome-wide redistribution of H3K27me3 is linked to genotoxic stress and defective growth.Proc Natl Acad Sci U S A. 2015 Nov 17;112(46):E6339-48. doi: 10.1073/pnas.1511377112. Epub 2015 Nov 2. Proc Natl Acad Sci U S A. 2015. PMID: 26578794 Free PMC article.
-
Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3.Genome Res. 2016 Aug;26(8):1069-80. doi: 10.1101/gr.203182.115. Epub 2016 Jun 3. Genome Res. 2016. PMID: 27260477 Free PMC article.
-
Neurospora importin α is required for normal heterochromatic formation and DNA methylation.PLoS Genet. 2015 Mar 20;11(3):e1005083. doi: 10.1371/journal.pgen.1005083. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25793375 Free PMC article.
-
Limited DNA Repair Gene Repertoire in Ascomycete Yeast Revealed by Comparative Genomics.Genome Biol Evol. 2019 Dec 1;11(12):3409-3423. doi: 10.1093/gbe/evz242. Genome Biol Evol. 2019. PMID: 31693105 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

