Role of ARF6, Rab11 and external Hsp90 in the trafficking and recycling of recombinant-soluble Neisseria meningitidis adhesin A (rNadA) in human epithelial cells
- PMID: 25347845
- PMCID: PMC4210143
- DOI: 10.1371/journal.pone.0110047
Role of ARF6, Rab11 and external Hsp90 in the trafficking and recycling of recombinant-soluble Neisseria meningitidis adhesin A (rNadA) in human epithelial cells
Abstract
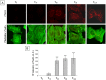
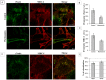
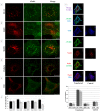
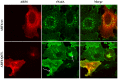
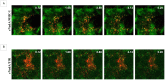
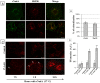
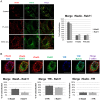
Neisseria meningitidis adhesin A (NadA) is a meningococcus surface protein thought to assist in the adhesion of the bacterium to host cells. We have previously shown that NadA also promotes bacterial internalization in a heterologous expression system. Here we have used the soluble recombinant NadA (rNadA) lacking the membrane anchor region to characterize its internalization route in Chang epithelial cells. Added to the culture medium, rNadA internalizes through a PI3K-dependent endocytosis process not mediated by the canonical clathrin or caveolin scaffolds, but instead follows an ARF6-regulated recycling pathway previously described for MHC-I. The intracellular pool of rNadA reaches a steady state level within one hour of incubation and colocalizes in endocytic vesicles with MHC-I and with the extracellularly labeled chaperone Hsp90. Treatment with membrane permeated and impermeable Hsp90 inhibitors 17-AAG and FITC-GA respectively, lead to intracellular accumulation of rNadA, strongly suggesting that the extracellular secreted pool of the chaperone is involved in rNadA intracellular trafficking. A significant number of intracellular vesicles containing rNadA recruit Rab11, a small GTPase associated to recycling endosomes, but do not contain transferrin receptor (TfR). Interestingly, cell treatment with Hsp90 inhibitors, including the membrane-impermeable FITC-GA, abolished Rab11-rNadA colocalization but do not interfere with Rab11-TfR colocalization. Collectively, these results are consistent with a model whereby rNadA internalizes into human epithelial cells hijacking the recycling endosome pathway and recycle back to the surface of the cell via an ARF6-dependent, Rab11 associated and Hsp90-regulated mechanism. The present study addresses for the first time a meningoccoccal adhesin mechanism of endocytosis and suggests a possible entry pathway engaged by N. meningitidis in primary infection of human epithelial cells.
Conflict of interest statement
Figures


Similar articles
-
Human heat shock protein (Hsp) 90 interferes with Neisseria meningitidis adhesin A (NadA)-mediated adhesion and invasion.Cell Microbiol. 2012 Mar;14(3):368-85. doi: 10.1111/j.1462-5822.2011.01722.x. Epub 2011 Dec 8. Cell Microbiol. 2012. PMID: 22066472
-
The soluble recombinant Neisseria meningitidis adhesin NadA(Δ351-405) stimulates human monocytes by binding to extracellular Hsp90.PLoS One. 2011;6(9):e25089. doi: 10.1371/journal.pone.0025089. Epub 2011 Sep 16. PLoS One. 2011. PMID: 21949862 Free PMC article.
-
Neisserial adhesin A (NadA) binds human Siglec-5 and Siglec-14 with high affinity and promotes bacterial adhesion/invasion.mBio. 2024 Aug 14;15(8):e0110724. doi: 10.1128/mbio.01107-24. Epub 2024 Jul 23. mBio. 2024. PMID: 39041817 Free PMC article.
-
Polarized endocytic transport: the roles of Rab11 and Rab11-FIPs in regulating cell polarity.Histol Histopathol. 2009 Sep;24(9):1171-80. doi: 10.14670/HH-24.1171. Histol Histopathol. 2009. PMID: 19609864 Free PMC article. Review.
-
Recycling Endosomes and Viral Infection.Viruses. 2016 Mar 8;8(3):64. doi: 10.3390/v8030064. Viruses. 2016. PMID: 27005655 Free PMC article. Review.
Cited by
-
How the Knowledge of Interactions between Meningococcus and the Human Immune System Has Been Used to Prepare Effective Neisseria meningitidis Vaccines.J Immunol Res. 2015;2015:189153. doi: 10.1155/2015/189153. Epub 2015 Aug 17. J Immunol Res. 2015. PMID: 26351643 Free PMC article. Review.
-
ARF1 and ARF6 regulate recycling of GRASP/Tamalin and the Rac1-GEF Dock180 during HGF-induced Rac1 activation.Small GTPases. 2018 May 4;9(3):242-259. doi: 10.1080/21541248.2016.1219186. Epub 2016 Aug 26. Small GTPases. 2018. PMID: 27562622 Free PMC article.
-
NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.mBio. 2018 Oct 16;9(5):e01914-18. doi: 10.1128/mBio.01914-18. mBio. 2018. PMID: 30327444 Free PMC article.
-
Heat Shock Protein 90 regulates encystation in Entamoeba.Front Microbiol. 2015 Oct 13;6:1125. doi: 10.3389/fmicb.2015.01125. eCollection 2015. Front Microbiol. 2015. PMID: 26528271 Free PMC article.
-
Regulation of Protein Transport Pathways by the Cytosolic Hsp90s.Biomolecules. 2022 Aug 5;12(8):1077. doi: 10.3390/biom12081077. Biomolecules. 2022. PMID: 36008972 Free PMC article. Review.
References
-
- Emonts M, Hazelzet JA, de Groot R, Hermans PW (2003) Host genetic determinants of Neisseria meningitidis infections. Lancet Infect Dis 3: 565–577. - PubMed
-
- Virji M (2009) Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat Rev Microbiol 7: 274–286. - PubMed
-
- Carbonnelle E, Hill DJ, Morand P, Griffiths NJ, Bourdoulous S, et al. (2009) Meningococcal interactions with the host. Vaccine 27 Suppl 2: B78–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

