Control of mammalian retrotransposons by cellular RNA processing activities
- PMID: 25346866
- PMCID: PMC4203495
- DOI: 10.4161/mge.28439
Control of mammalian retrotransposons by cellular RNA processing activities
Abstract
Retrotransposons make up roughly 50% of the mammalian genome and have played an important role in genome evolution. A small fraction of non-LTR retrotransposons, LINE-1 and SINE elements, is currently active in the human genome. These elements move in our genome using an intermediate RNA and a reverse transcriptase activity by a copy and paste mechanism. Their ongoing mobilization can impact the human genome leading to several human disorders. However, how the cell controls the activity of these elements minimizing their mutagenic effect is not fully understood. Recent studies have highlighted that the intermediate RNA of retrotransposons is a target of different mechanisms that limit the mobilization of endogenous retrotransposons in mammals. Here, we provide an overview of recent discoveries that show how RNA processing events can act to control the activity of mammalian retrotransposons and discuss several arising questions that remain to be answered.
Keywords: DGCR8; Dicer; Drosha; LINE-1; Microprocessor; SINE-1; microRNAs; retrotransposon; small RNAs; transposable elements.
Figures
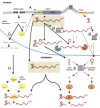
Comment on
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Cáceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20:1173–81. doi: 10.1038/nsmb.2658.
Similar articles
-
Control of chicken CR1 retrotransposons is independent of Dicer-mediated RNA interference pathway.BMC Biol. 2009 Aug 19;7:53. doi: 10.1186/1741-7007-7-53. BMC Biol. 2009. PMID: 19691826 Free PMC article.
-
The Microprocessor controls the activity of mammalian retrotransposons.Nat Struct Mol Biol. 2013 Oct;20(10):1173-81. doi: 10.1038/nsmb.2658. Epub 2013 Sep 1. Nat Struct Mol Biol. 2013. PMID: 23995758 Free PMC article.
-
The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0061-2014. doi: 10.1128/microbiolspec.MDNA3-0061-2014. Microbiol Spectr. 2015. PMID: 26104698 Free PMC article. Review.
-
LINE-1 Retrotransposition Assays in Embryonic Stem Cells.Methods Mol Biol. 2023;2607:257-309. doi: 10.1007/978-1-0716-2883-6_13. Methods Mol Biol. 2023. PMID: 36449167
-
Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health.Genome Res. 2008 Mar;18(3):343-58. doi: 10.1101/gr.5558208. Epub 2008 Feb 6. Genome Res. 2008. PMID: 18256243 Review.
Cited by
-
Restricting retrotransposons: a review.Mob DNA. 2016 Aug 11;7:16. doi: 10.1186/s13100-016-0070-z. eCollection 2016. Mob DNA. 2016. PMID: 27525044 Free PMC article. Review.
-
Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci.Elife. 2016 Mar 26;5:e13926. doi: 10.7554/eLife.13926. Elife. 2016. PMID: 27016617 Free PMC article.
-
Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil.Mol Biol Evol. 2015 May;32(5):1268-83. doi: 10.1093/molbev/msv017. Epub 2015 Jan 28. Mol Biol Evol. 2015. PMID: 25633377 Free PMC article.
-
RNase H2, mutated in Aicardi-Goutières syndrome, promotes LINE-1 retrotransposition.EMBO J. 2018 Aug 1;37(15):e98506. doi: 10.15252/embj.201798506. Epub 2018 Jun 29. EMBO J. 2018. PMID: 29959219 Free PMC article.
-
Navigating the brain and aging: exploring the impact of transposable elements from health to disease.Front Cell Dev Biol. 2024 Feb 27;12:1357576. doi: 10.3389/fcell.2024.1357576. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38476259 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
