Design of gem-difluoro-bis-tetrahydrofuran as P2 ligand for HIV-1 protease inhibitors to improve brain penetration: synthesis, X-ray studies, and biological evaluation
- PMID: 25336073
- PMCID: PMC4419590
- DOI: 10.1002/cmdc.201402358
Design of gem-difluoro-bis-tetrahydrofuran as P2 ligand for HIV-1 protease inhibitors to improve brain penetration: synthesis, X-ray studies, and biological evaluation
Abstract
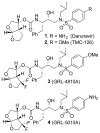
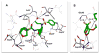
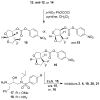
The structure-based design, synthesis, biological evaluation, and X-ray structural studies of fluorine-containing HIV-1 protease inhibitors are described. The synthesis of both enantiomers of the gem-difluoro-bis-THF ligands was carried out in a stereoselective manner using a Reformatskii-Claisen reaction as the key step. Optically active ligands were converted into protease inhibitors. Two of these inhibitors, (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl(2S,3R)-3-hydroxy-4-((N-isobutyl-4-methoxyphenyl)sulfonamido)-1-phenylbutan-2-yl) carbamate (3) and (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl(2S,3R)-3-hydroxy-4-((N-isobutyl-4-aminophenyl)sulfonamido)phenylbutan-2-yl) carbamate (4), exhibited HIV-1 protease inhibitory Ki values in the picomolar range. Both 3 and 4 showed very potent antiviral activity, with respective EC50 values of 0.8 and 3.1 nM against the laboratory strain HIV-1LAI . The two inhibitors exhibited better lipophilicity profiles than darunavir, and also showed much improved blood-brain barrier permeability in an in vitro model. A high-resolution X-ray structure of inhibitor 4 in complex with HIV-1 protease was determined, revealing that the fluorinated ligand makes extensive interactions with the S2 subsite of HIV-1 protease, including hydrogen bonding interactions with the protease backbone atoms. Moreover, both fluorine atoms on the bis-THF ligand formed strong interactions with the flap Gly 48 carbonyl oxygen atom.
Keywords: HIV-1 protease; antiviral agents; blood-brain barrier; inhibitors; ligands; multidrug resistance.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
Similar articles
-
Nonpeptidal P2 ligands for HIV protease inhibitors: structure-based design, synthesis, and biological evaluation.J Med Chem. 1996 Aug 16;39(17):3278-90. doi: 10.1021/jm960128k. J Med Chem. 1996. PMID: 8765511
-
Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.J Med Chem. 2015 Sep 10;58(17):6994-7006. doi: 10.1021/acs.jmedchem.5b00900. Epub 2015 Aug 25. J Med Chem. 2015. PMID: 26306007 Free PMC article.
-
Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.Org Biomol Chem. 2024 Sep 18;22(36):7354-7372. doi: 10.1039/d4ob00506f. Org Biomol Chem. 2024. PMID: 38973505
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
Highly resistant HIV-1 proteases and strategies for their inhibition.Future Med Chem. 2015;7(8):1023-38. doi: 10.4155/fmc.15.44. Future Med Chem. 2015. PMID: 26062399 Free PMC article. Review.
-
Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS.J Med Chem. 2016 Jun 9;59(11):5172-208. doi: 10.1021/acs.jmedchem.5b01697. Epub 2016 Jan 22. J Med Chem. 2016. PMID: 26799988 Free PMC article. Review.
-
Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance.Chem Commun (Camb). 2022 Oct 20;58(84):11762-11782. doi: 10.1039/d2cc04541a. Chem Commun (Camb). 2022. PMID: 36200462 Free PMC article. Review.
-
Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.Antimicrob Agents Chemother. 2022 Feb 15;66(2):e0171521. doi: 10.1128/AAC.01715-21. Epub 2022 Jan 3. Antimicrob Agents Chemother. 2022. PMID: 34978889 Free PMC article.
-
Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.ChemMedChem. 2018 Apr 23;13(8):803-815. doi: 10.1002/cmdc.201700824. Epub 2018 Mar 15. ChemMedChem. 2018. PMID: 29437300 Free PMC article.
References
-
- Purser S, Moore PR, Swallow S, Gouverneur V. Chem Soc Rev. 2008;37:320–330. - PubMed
-
- Bohm HJ, Banner D, Bendels S, Kansy M, Kuhn B, Muller K, Obst-Sander U, Stahl M. Chembiochem. 2004;5:637–643. - PubMed
-
- Park BK, Kitteringham NR, O’Neill PM. Annu Rev Pharmacol Toxicol. 2001;41:443–470. - PubMed
-
- Smart BE. J Fluorine Chem. 2001;109:3–11.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

