Polycomb Repressive Complex 2 regulates lineage fidelity during embryonic stem cell differentiation
- PMID: 25333635
- PMCID: PMC4204901
- DOI: 10.1371/journal.pone.0110498
Polycomb Repressive Complex 2 regulates lineage fidelity during embryonic stem cell differentiation
Abstract
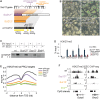
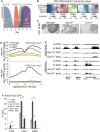
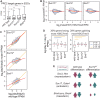
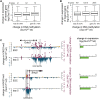
Polycomb Repressive Complex 2 (PRC2) catalyzes histone H3 lysine 27 tri-methylation (H3K27me3), an epigenetic modification associated with gene repression. H3K27me3 is enriched at the promoters of a large cohort of developmental genes in embryonic stem cells (ESCs). Loss of H3K27me3 leads to a failure of ESCs to properly differentiate, making it difficult to determine the precise roles of PRC2 during lineage commitment. Moreover, while studies suggest that PRC2 prevents DNA methylation, how these two epigenetic regulators coordinate to regulate lineage programs is poorly understood. Using several PRC2 mutant ESC lines that maintain varying levels of H3K27me3, we found that partial maintenance of H3K27me3 allowed for proper temporal activation of lineage genes during directed differentiation of ESCs to spinal motor neurons (SMNs). In contrast, genes that function to specify other lineages failed to be repressed in these cells, suggesting that PRC2 is also necessary for lineage fidelity. We also found that loss of H3K27me3 leads to a modest gain in DNA methylation at PRC2 target regions in both ESCs and in SMNs. Our study demonstrates a critical role for PRC2 in safeguarding lineage decisions and in protecting genes against inappropriate DNA methylation.
Conflict of interest statement
Figures

Similar articles
-
Polycomb protein EED is required for silencing of pluripotency genes upon ESC differentiation.Stem Cell Rev Rep. 2015 Feb;11(1):50-61. doi: 10.1007/s12015-014-9550-z. Stem Cell Rev Rep. 2015. PMID: 25134795
-
Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation.Mol Cell. 2015 Mar 5;57(5):769-783. doi: 10.1016/j.molcel.2014.12.020. Epub 2015 Jan 22. Mol Cell. 2015. PMID: 25620564 Free PMC article.
-
Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells.Genome Biol. 2013 Aug 29;14(8):R91. doi: 10.1186/gb-2013-14-8-r91. Genome Biol. 2013. PMID: 23987249 Free PMC article.
-
Epigenetic regulation of embryonic ectoderm development in stem cell differentiation and transformation during ontogenesis.Cell Prolif. 2023 Apr;56(4):e13413. doi: 10.1111/cpr.13413. Epub 2023 Feb 1. Cell Prolif. 2023. PMID: 36727213 Free PMC article. Review.
-
The Complexity of PRC2 Subcomplexes.Trends Cell Biol. 2019 Aug;29(8):660-671. doi: 10.1016/j.tcb.2019.05.004. Epub 2019 Jun 6. Trends Cell Biol. 2019. PMID: 31178244 Review.
Cited by
-
Re-expression of miR-200s in claudin-low mammary tumor cells alters cell shape and reduces proliferation and invasion potentially through modulating other miRNAs and SUZ12 regulated genes.Cancer Cell Int. 2021 Feb 4;21(1):89. doi: 10.1186/s12935-021-01784-4. Cancer Cell Int. 2021. PMID: 33541373 Free PMC article.
-
The role of LINC01419 in regulating the cell stemness in lung adenocarcinoma through recruiting EZH2 and regulating FBP1 expression.Biol Direct. 2022 Sep 1;17(1):23. doi: 10.1186/s13062-022-00336-8. Biol Direct. 2022. PMID: 36050791 Free PMC article.
-
EpCAM-regulated intramembrane proteolysis induces a cancer stem cell-like gene signature in hepatitis B virus-infected hepatocytes.J Hepatol. 2016 Nov;65(5):888-898. doi: 10.1016/j.jhep.2016.05.022. Epub 2016 May 26. J Hepatol. 2016. PMID: 27238755 Free PMC article.
-
The nucleosome remodeling and deacetylase complex protein CHD4 regulates neural differentiation of mouse embryonic stem cells by down-regulating p53.J Biol Chem. 2019 Jan 4;294(1):195-209. doi: 10.1074/jbc.RA118.004086. Epub 2018 Nov 8. J Biol Chem. 2019. PMID: 30409903 Free PMC article.
-
Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity.Elife. 2018 Oct 12;7:e38080. doi: 10.7554/eLife.38080. Elife. 2018. PMID: 30311912 Free PMC article.
References
-
- Faust C, Schumacher A, Holdener B, Magnuson T (1995) The eed mutation disrupts anterior mesoderm production in mice. Development 121: 273–285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

