Picosecond-resolved fluorescence studies of substrate and cofactor-binding domain mutants in a thermophilic alcohol dehydrogenase uncover an extended network of communication
- PMID: 25314615
- PMCID: PMC4210157
- DOI: 10.1021/ja506667k
Picosecond-resolved fluorescence studies of substrate and cofactor-binding domain mutants in a thermophilic alcohol dehydrogenase uncover an extended network of communication
Abstract
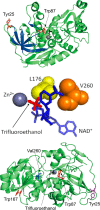
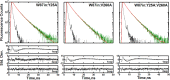
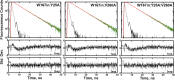
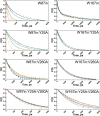
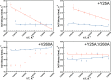
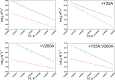
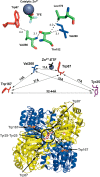
Time-resolved fluorescence dynamics are investigated in two mutants of a thermophilic alcohol dehydrogenase (ht-ADH): Y25A (at the dimer interface) and V260A (at the cofactor-binding domain). These residues, ca. 32 Å apart, are shown to exhibit opposing low-temperature effects on the hydride tunneling step. Using single-tryptophan constructs at the active site (Trp87) and a remote, surface-exposed site (Trp167), time-dependent Stokes shifts and collisional quenching data allow an analysis of intra-protein dynamical communication. A double mutant, Y25A:V260A, was also inserted into each single-Trp construct and analyzed accordingly. None of the mutations affect fluorescence lifetimes, Stokes shift relaxation rates, and quenching data for the surface-exposed Trp167 to an appreciable extent. By contrast, fluorescent probes of the active-site tryptophan 87 reveal distinctive forms of dynamical communication. Stokes shifts show that the distal Y25A increases active-site flexibility, V260A introduces a temperature-dependent equilibration process not previously reported by such measurements, and the double mutant (Y25A:V260A) eliminates the temperature-dependent transition sensed by the active-site tryptophan in the presence of V260A. Collisional quenching data at Trp87 further show a structural change in the active-site environment/solvation for V260A. In the aggregate, the temperature dependencies of the fluorescence data are distinct from the breaks in behavior previously reported for catalysis and hydrogen/deuterium exchange, attributed to time scales for the interconversion of protein conformational substates that are slower and more global than the local motions monitored within. An extended network of dynamical communication between the protein dimer surface and substrate- and cofactor-binding domains emerges from the flourescent data.
Figures
Similar articles
-
Temperature-Jump Fluorescence Provides Evidence for Fully Reversible Microsecond Dynamics in a Thermophilic Alcohol Dehydrogenase.J Am Chem Soc. 2015 Aug 19;137(32):10060-3. doi: 10.1021/jacs.5b04413. Epub 2015 Aug 10. J Am Chem Soc. 2015. PMID: 26223665 Free PMC article.
-
Picosecond-resolved fluorescent probes at functionally distinct tryptophans within a thermophilic alcohol dehydrogenase: relationship of temperature-dependent changes in fluorescence to catalysis.J Phys Chem B. 2014 Jun 12;118(23):6049-61. doi: 10.1021/jp500825x. Epub 2014 Jun 3. J Phys Chem B. 2014. PMID: 24892947 Free PMC article.
-
Identification of a long-range protein network that modulates active site dynamics in extremophilic alcohol dehydrogenases.J Biol Chem. 2013 May 17;288(20):14087-14097. doi: 10.1074/jbc.M113.453951. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525111 Free PMC article.
-
Dynamically achieved active site precision in enzyme catalysis.Acc Chem Res. 2015 Feb 17;48(2):449-56. doi: 10.1021/ar5003347. Epub 2014 Dec 24. Acc Chem Res. 2015. PMID: 25539048 Free PMC article. Review.
-
Conformational changes and catalysis by alcohol dehydrogenase.Arch Biochem Biophys. 2010 Jan 1;493(1):3-12. doi: 10.1016/j.abb.2009.07.001. Epub 2009 Jul 5. Arch Biochem Biophys. 2010. PMID: 19583966 Free PMC article. Review.
Cited by
-
Detecting and Characterizing the Kinetic Activation of Thermal Networks in Proteins: Thermal Transfer from a Distal, Solvent-Exposed Loop to the Active Site in Soybean Lipoxygenase.J Phys Chem B. 2019 Oct 17;123(41):8662-8674. doi: 10.1021/acs.jpcb.9b07228. Epub 2019 Oct 3. J Phys Chem B. 2019. PMID: 31580070 Free PMC article.
-
Isotope Substitution of Promiscuous Alcohol Dehydrogenase Reveals the Origin of Substrate Preference in the Transition State.Angew Chem Int Ed Engl. 2018 Mar 12;57(12):3128-3131. doi: 10.1002/anie.201712826. Epub 2018 Feb 19. Angew Chem Int Ed Engl. 2018. PMID: 29341402 Free PMC article.
-
Temperature-Jump Fluorescence Provides Evidence for Fully Reversible Microsecond Dynamics in a Thermophilic Alcohol Dehydrogenase.J Am Chem Soc. 2015 Aug 19;137(32):10060-3. doi: 10.1021/jacs.5b04413. Epub 2015 Aug 10. J Am Chem Soc. 2015. PMID: 26223665 Free PMC article.
-
Linking protein motion to enzyme catalysis.Molecules. 2015 Jan 13;20(1):1192-209. doi: 10.3390/molecules20011192. Molecules. 2015. PMID: 25591120 Free PMC article. Review.
-
Hydrogen deuterium exchange defines catalytically linked regions of protein flexibility in the catechol O-methyltransferase reaction.Proc Natl Acad Sci U S A. 2020 May 19;117(20):10797-10805. doi: 10.1073/pnas.1917219117. Epub 2020 May 5. Proc Natl Acad Sci U S A. 2020. PMID: 32371482 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

