Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome
- PMID: 25313070
- PMCID: PMC4246342
- DOI: 10.1073/pnas.1418000111
Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome
Abstract
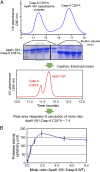
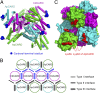
Autocatalytic activation of an initiator caspase triggers the onset of apoptosis. In dying cells, caspase-9 activation is mediated by a multimeric adaptor complex known as the Apaf-1 apoptosome. The molecular mechanism by which caspase-9 is activated by the Apaf-1 apoptosome remains largely unknown. Here we demonstrate that the previously reported 1:1 interaction between Apaf-1 caspase recruitment domain (CARD) and caspase-9 CARD is insufficient for the activation of caspase-9. Rather, formation of a multimeric CARD:CARD assembly between Apaf-1 and caspase-9, which requires three types of distinct interfaces, underlies caspase-9 activation. Importantly, an additional surface area on the multimeric CARD assembly is essential for caspase-9 activation. Together, these findings reveal mechanistic insights into the activation of caspase-9 by the Apaf-1 apoptosome and support the induced conformation model for initiator caspase activation by adaptor complexes.
Keywords: CARD; apoptosis; caspase activation; induced proximity; mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.Genes Dev. 2015 Feb 1;29(3):277-87. doi: 10.1101/gad.255877.114. Genes Dev. 2015. PMID: 25644603 Free PMC article.
-
Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1542-1547. doi: 10.1073/pnas.1620626114. Epub 2017 Jan 31. Proc Natl Acad Sci U S A. 2017. PMID: 28143931 Free PMC article.
-
Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.Structure. 2017 Mar 7;25(3):407-420. doi: 10.1016/j.str.2016.12.019. Epub 2017 Jan 19. Structure. 2017. PMID: 28111022
-
Apoptosome structure, assembly, and procaspase activation.Structure. 2013 Apr 2;21(4):501-15. doi: 10.1016/j.str.2013.02.024. Structure. 2013. PMID: 23561633 Free PMC article. Review.
-
Apoptosome: a platform for the activation of initiator caspases.Cell Death Differ. 2007 Jan;14(1):56-65. doi: 10.1038/sj.cdd.4402028. Epub 2006 Sep 15. Cell Death Differ. 2007. PMID: 16977332 Review.
Cited by
-
Inactivated Tianjin strain, a novel genotype of Sendai virus, induces apoptosis in HeLa, NCI-H446 and Hep3B cells.Oncol Lett. 2016 Jul;12(1):49-56. doi: 10.3892/ol.2016.4570. Epub 2016 May 13. Oncol Lett. 2016. PMID: 27347098 Free PMC article.
-
Activation of caspase-9 on the apoptosome as studied by methyl-TROSY NMR.Proc Natl Acad Sci U S A. 2023 Dec 19;120(51):e2310944120. doi: 10.1073/pnas.2310944120. Epub 2023 Dec 12. Proc Natl Acad Sci U S A. 2023. PMID: 38085782 Free PMC article.
-
Ivabradine Treatment Reduces Cardiomyocyte Apoptosis in a Murine Model of Chronic Viral Myocarditis.Front Pharmacol. 2018 Mar 5;9:182. doi: 10.3389/fphar.2018.00182. eCollection 2018. Front Pharmacol. 2018. PMID: 29556195 Free PMC article.
-
Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018.Cell Death Differ. 2018 Mar;25(3):486-541. doi: 10.1038/s41418-017-0012-4. Epub 2018 Jan 23. Cell Death Differ. 2018. PMID: 29362479 Free PMC article. Review.
-
The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold.Sci Rep. 2016 Nov 23;6:37267. doi: 10.1038/srep37267. Sci Rep. 2016. PMID: 27876844 Free PMC article.
References
-
- Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281(5381):1312–1316. - PubMed
-
- Horvitz HR. Worms, life, and death (Nobel lecture) ChemBioChem. 2003;4(8):697–711. - PubMed
-
- Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116(2):205–219. - PubMed
-
- Yan N, Shi Y. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol. 2005;21:35–56. - PubMed
-
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5(11):897–907. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

