Fragmentation follows structure: top-down mass spectrometry elucidates the topology of engineered cystine-knot miniproteins
- PMID: 25303319
- PMCID: PMC4193770
- DOI: 10.1371/journal.pone.0108626
Fragmentation follows structure: top-down mass spectrometry elucidates the topology of engineered cystine-knot miniproteins
Abstract
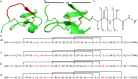
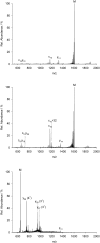
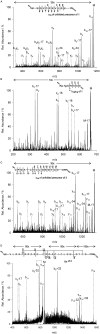
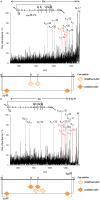
Over the last decades the field of pharmaceutically relevant peptides has enormously expanded. Among them, several peptide families exist that contain three or more disulfide bonds. In this context, elucidation of the disulfide patterns is extremely important as these motifs are often prerequisites for folding, stability, and activity. An example of this structure-determining pattern is a cystine knot which comprises three constrained disulfide bonds and represents a core element in a vast number of mechanically interlocked peptidic structures possessing different biological activities. Herein, we present our studies on disulfide pattern determination and structure elucidation of cystine-knot miniproteins derived from Momordica cochinchinensis peptide MCoTI-II, which act as potent inhibitors of human matriptase-1. A top-down mass spectrometric analysis of the oxidised and bioactive peptides is described. Following the detailed sequencing of the peptide backbone, interpretation of the MS(3) spectra allowed for the verification of the knotted topology of the examined miniproteins. Moreover, we found that the fragmentation pattern depends on the knottin's folding state, hence, tertiary structure, which to our knowledge has not been described for a top-down MS approach before.
Conflict of interest statement
Figures
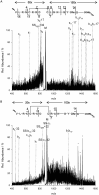
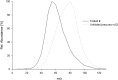
Similar articles
-
Knots in rings. The circular knotted protein Momordica cochinchinensis trypsin inhibitor-II folds via a stable two-disulfide intermediate.J Biol Chem. 2006 Mar 24;281(12):8224-32. doi: 10.1074/jbc.M513399200. Epub 2006 Jan 23. J Biol Chem. 2006. PMID: 16547012
-
Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides.J Biol Chem. 2003 Feb 21;278(8):6314-22. doi: 10.1074/jbc.M210492200. Epub 2002 Dec 12. J Biol Chem. 2003. PMID: 12482862
-
Computational analysis of the MCoTI-II plant defence knottin reveals a novel intermediate conformation that facilitates trypsin binding.Sci Rep. 2016 Mar 15;6:23174. doi: 10.1038/srep23174. Sci Rep. 2016. PMID: 26975976 Free PMC article.
-
The cyclotide family of circular miniproteins: nature's combinatorial peptide template.Biopolymers. 2006;84(3):250-66. doi: 10.1002/bip.20451. Biopolymers. 2006. PMID: 16440288 Review.
-
Oxidative folding of the cystine knot motif in cyclotide proteins.Protein Pept Lett. 2005 Feb;12(2):147-52. doi: 10.2174/0929866053005863. Protein Pept Lett. 2005. PMID: 15723640 Review.
Cited by
-
Proteomic Approaches to Study Cysteine Oxidation: Applications in Neurodegenerative Diseases.Front Mol Neurosci. 2021 Jun 9;14:678837. doi: 10.3389/fnmol.2021.678837. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34177463 Free PMC article. Review.
References
-
- Chen J, Shiyanov P, Schlager JJ, Green KB (2012) A pseudo MS3 approach for identification of disulfide-bonded proteins: uncommon product ions and database search. J Am Soc Mass Spectrom 23: 225–243. - PubMed
-
- Seidler J, Zinn N, Boehm ME, Lehmann WD (2010) De novo sequencing of peptides by MS/MS. Proteomics 10: 634–649. - PubMed
-
- Goransson U, Craik DJ (2003) Disulfide mapping of the cyclotide kalata B1. Chemical proof of the cystic cystine knot motif. J Biol Chem 278: 48188–48196. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

