Current Insights into Research on Rice stripe virus
- PMID: 25288949
- PMCID: PMC4174810
- DOI: 10.5423/PPJ.RW.10.2012.0158
Current Insights into Research on Rice stripe virus
Abstract
Rice stripe virus (RSV) is one of the most destructive viruses of rice, and greatly reduces rice production in China, Japan, and Korea, where mostly japonica cultivars of rice are grown. RSV is transmitted by the small brown plant-hopper (SBPH) in a persistent and circulative-propagative manner. Several methods have been developed for detection of RSV, which is composed of four single-stranded RNAs that encode seven proteins. Genome sequence data and comparative phylogenetic analysis have been used to identify the origin and diversity of RSV isolates. Several rice varieties resistant to RSV have been selected and QTL analysis and fine mapping have been intensively performed to map RSV resistance loci or genes. RSV genes have been used to generate several genetically modified transgenic rice plants with RSV resistance. Recently, genome-wide transcriptome analyses and deep sequencing have been used to identify mRNAs and small RNAs involved in RSV infection; several rice host factors that interact with RSV proteins have also been identified. In this article, we review the current statues of RSV research and propose integrated approaches for the study of interactions among RSV, rice, and the SBPH.
Keywords: Rice stripe virus; genome; quantitative trait locus; resistance; rice.
Figures
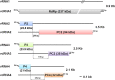
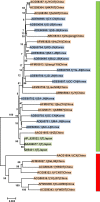
Similar articles
-
NS3 Protein from Rice stripe virus affects the expression of endogenous genes in Nicotiana benthamiana.Virol J. 2018 Jun 25;15(1):105. doi: 10.1186/s12985-018-1014-7. Virol J. 2018. PMID: 29940994 Free PMC article.
-
Detection of quantitative trait loci (QTLs) for resistances to small brown planthopper and rice stripe virus in rice using recombinant inbred lines.Int J Mol Sci. 2013 Apr 16;14(4):8406-21. doi: 10.3390/ijms14048406. Int J Mol Sci. 2013. PMID: 23591851 Free PMC article.
-
Suppression of NS3 and MP is important for the stable inheritance of RNAi-mediated rice stripe virus (RSV) resistance obtained by targeting the fully complementary RSV-CP gene.Mol Cells. 2012 Jan;33(1):43-51. doi: 10.1007/s10059-012-2185-5. Epub 2011 Nov 29. Mol Cells. 2012. PMID: 22134721 Free PMC article.
-
Organ-specific transcriptome response of the small brown planthopper toward rice stripe virus.Insect Biochem Mol Biol. 2016 Mar;70:60-72. doi: 10.1016/j.ibmb.2015.11.009. Epub 2015 Dec 8. Insect Biochem Mol Biol. 2016. PMID: 26678499
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
Cited by
-
Genetic variations and gene expression profiles of Rice Black-streaked dwarf virus (RBSDV) in different host plants and insect vectors: insights from RNA-Seq analysis.BMC Genomics. 2024 Jul 30;25(1):736. doi: 10.1186/s12864-024-10649-9. BMC Genomics. 2024. PMID: 39080552 Free PMC article.
-
Development of Rice Stripe Tenuivirus Minireplicon Reverse Genetics Systems Suitable for Analyses of Viral Replication and Intercellular Movement.Front Microbiol. 2021 Mar 23;12:655256. doi: 10.3389/fmicb.2021.655256. eCollection 2021. Front Microbiol. 2021. PMID: 33833749 Free PMC article.
-
Rice stripe virus p2 Colocalizes and Interacts with Arabidopsis Cajal Bodies and Its Domains in Plant Cells.Biomed Res Int. 2020 Jun 16;2020:5182164. doi: 10.1155/2020/5182164. eCollection 2020. Biomed Res Int. 2020. Retraction in: Biomed Res Int. 2024 Mar 20;2024:9857937. doi: 10.1155/2024/9857937. PMID: 32685498 Free PMC article. Retracted.
-
NS3 Protein from Rice stripe virus affects the expression of endogenous genes in Nicotiana benthamiana.Virol J. 2018 Jun 25;15(1):105. doi: 10.1186/s12985-018-1014-7. Virol J. 2018. PMID: 29940994 Free PMC article.
-
Rice stripe virus nonstructural protein 3 suppresses plant defence responses mediated by the MEL-SHMT1 module.Mol Plant Pathol. 2023 Nov;24(11):1359-1369. doi: 10.1111/mpp.13373. Epub 2023 Jul 5. Mol Plant Pathol. 2023. PMID: 37404045 Free PMC article.
References
-
- Abo ME, Sy AA. Rice virus diseases: epidemiology and management strategies. J Sustain Agr. 1997;11:113–134.
-
- Cai L, Ma X, Lin K, Deng K, Zhao S, Li C. Detecting Rice stripe virus (RSV) in the small brown planthopper Laodelphax striatellus with high specificity by RT-PCR. J Virol Methods. 2003;112:115–120. - PubMed
-
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RWH. The capsnatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

