tBRD-1 selectively controls gene activity in the Drosophila testis and interacts with two new members of the bromodomain and extra-terminal (BET) family
- PMID: 25251222
- PMCID: PMC4177214
- DOI: 10.1371/journal.pone.0108267
tBRD-1 selectively controls gene activity in the Drosophila testis and interacts with two new members of the bromodomain and extra-terminal (BET) family
Abstract
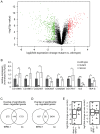
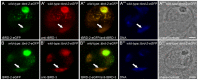
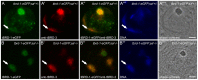
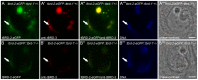
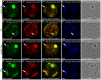
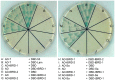
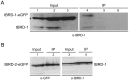
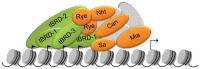
Multicellular organisms have evolved specialized mechanisms to control transcription in a spatial and temporal manner. Gene activation is tightly linked to histone acetylation on lysine residues that can be recognized by bromodomains. Previously, the testis-specifically expressed bromodomain protein tBRD-1 was identified in Drosophila. Expression of tBRD-1 is restricted to highly transcriptionally active primary spermatocytes. tBRD-1 is essential for male fertility and proposed to act as a co-factor of testis-specific TATA box binding protein-associated factors (tTAFs) for testis-specific transcription. Here, we performed microarray analyses to compare the transcriptomes of tbrd-1 mutant testes and wild-type testes. Our data confirmed that tBRD-1 controls gene activity in male germ cells. Additionally, comparing the transcriptomes of tbrd-1 and tTAF mutant testes revealed a subset of common target genes. We also characterized two new members of the bromodomain and extra-terminal (BET) family, tBRD-2 and tBRD-3. In contrast to other members of the BET family in animals, both possess only a single bromodomain, a characteristic feature of plant BET family members. Immunohistology techniques not only revealed that tBRD-2 and tBRD-3 partially co-localize with tBRD-1 and tTAFs in primary spermatocytes, but also that their proper subcellular distribution was impaired in tbrd-1 and tTAF mutant testes. Treating cultured male germ cells with inhibitors showed that localization of tBRD-2 and tBRD-3 depends on the acetylation status within primary spermatocytes. Yeast two-hybrid assays and co-immunoprecipitations using fly testes protein extracts demonstrated that tBRD-1 is able to form homodimers as well as heterodimers with tBRD-2, tBRD-3, and tTAFs. These data reveal for the first time the existence of single bromodomain BET proteins in animals, as well as evidence for a complex containing tBRDs and tTAFs that regulates transcription of a subset of genes with relevance for spermiogenesis.
Conflict of interest statement
Figures

Similar articles
-
tBRD-1 and tBRD-2 regulate expression of genes necessary for spermatid differentiation.Biol Open. 2017 Apr 15;6(4):439-448. doi: 10.1242/bio.022467. Biol Open. 2017. PMID: 28235844 Free PMC article.
-
Two bromodomain proteins functionally interact to recapitulate an essential BRDT-like function in Drosophila spermatocytes.Open Biol. 2015 Feb;5(2):140145. doi: 10.1098/rsob.140145. Open Biol. 2015. PMID: 25652540 Free PMC article.
-
The bromodomain-containing protein tBRD-1 is specifically expressed in spermatocytes and is essential for male fertility.Biol Open. 2012 Jun 15;1(6):597-606. doi: 10.1242/bio.20121255. Epub 2012 May 9. Biol Open. 2012. PMID: 23213453 Free PMC article.
-
The role of the double bromodomain-containing BET genes during mammalian spermatogenesis.Curr Top Dev Biol. 2013;102:293-326. doi: 10.1016/B978-0-12-416024-8.00011-8. Curr Top Dev Biol. 2013. PMID: 23287038 Free PMC article. Review.
-
The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins.Int J Mol Sci. 2016 Nov 7;17(11):1849. doi: 10.3390/ijms17111849. Int J Mol Sci. 2016. PMID: 27827996 Free PMC article. Review.
Cited by
-
tBRD-1 and tBRD-2 regulate expression of genes necessary for spermatid differentiation.Biol Open. 2017 Apr 15;6(4):439-448. doi: 10.1242/bio.022467. Biol Open. 2017. PMID: 28235844 Free PMC article.
-
Nejire/dCBP-mediated histone H3 acetylation during spermatogenesis is essential for male fertility in Drosophila melanogaster.PLoS One. 2018 Sep 7;13(9):e0203622. doi: 10.1371/journal.pone.0203622. eCollection 2018. PLoS One. 2018. PMID: 30192860 Free PMC article.
-
Identification of genes functionally involved in the detrimental effects of mutant histone H3.3-K27M in Drosophila melanogaster.Neuro Oncol. 2019 May 6;21(5):628-639. doi: 10.1093/neuonc/noz021. Neuro Oncol. 2019. PMID: 30715493 Free PMC article.
-
Two bromodomain proteins functionally interact to recapitulate an essential BRDT-like function in Drosophila spermatocytes.Open Biol. 2015 Feb;5(2):140145. doi: 10.1098/rsob.140145. Open Biol. 2015. PMID: 25652540 Free PMC article.
-
Testes Proteases Expression and Hybrid Male Sterility Between Subspecies of Drosophila pseudoobscura.G3 (Bethesda). 2019 Apr 9;9(4):1065-1074. doi: 10.1534/g3.119.300580. G3 (Bethesda). 2019. PMID: 30723102 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

