Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells
- PMID: 25241740
- PMCID: PMC4266008
- DOI: 10.1016/j.stemcr.2014.07.003
Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells
Abstract
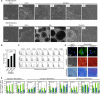
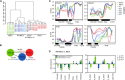
Standardization of mesenchymal stromal cells (MSCs) remains a major obstacle in regenerative medicine. Starting material and culture expansion affect cell preparations and render comparison between studies difficult. In contrast, induced pluripotent stem cells (iPSCs) assimilate toward a ground state and may therefore give rise to more standardized cell preparations. We reprogrammed MSCs into iPSCs, which were subsequently redifferentiated toward MSCs. These iPS-MSCs revealed similar morphology, immunophenotype, in vitro differentiation potential, and gene expression profiles as primary MSCs. However, iPS-MSCs were impaired in suppressing T cell proliferation. DNA methylation (DNAm) profiles of iPSCs maintained donor-specific characteristics, whereas tissue-specific, senescence-associated, and age-related DNAm patterns were erased during reprogramming. iPS-MSCs reacquired senescence-associated DNAm during culture expansion, but they remained rejuvenated with regard to age-related DNAm. Overall, iPS-MSCs are similar to MSCs, but they reveal incomplete reacquisition of immunomodulatory function and MSC-specific DNAm patterns-particularly of DNAm patterns associated with tissue type and aging.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Tracking of epigenetic changes during hematopoietic differentiation of induced pluripotent stem cells.Clin Epigenetics. 2019 Feb 4;11(1):19. doi: 10.1186/s13148-019-0617-1. Clin Epigenetics. 2019. PMID: 30717806 Free PMC article.
-
Interrupted reprogramming into induced pluripotent stem cells does not rejuvenate human mesenchymal stromal cells.Sci Rep. 2018 Aug 3;8(1):11676. doi: 10.1038/s41598-018-30069-6. Sci Rep. 2018. PMID: 30076334 Free PMC article.
-
Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells.Stem Cells Transl Med. 2012 Feb;1(2):83-95. doi: 10.5966/sctm.2011-0022. Epub 2012 Feb 7. Stem Cells Transl Med. 2012. PMID: 23197756 Free PMC article.
-
Production of Mesenchymal Stem Cells Through Stem Cell Reprogramming.Int J Mol Sci. 2019 Apr 18;20(8):1922. doi: 10.3390/ijms20081922. Int J Mol Sci. 2019. PMID: 31003536 Free PMC article. Review.
-
Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells.Int J Mol Sci. 2018 Mar 21;19(4):930. doi: 10.3390/ijms19040930. Int J Mol Sci. 2018. PMID: 29561778 Free PMC article. Review.
Cited by
-
No Time to Age: Uncoupling Aging from Chronological Time.Genes (Basel). 2021 Apr 21;12(5):611. doi: 10.3390/genes12050611. Genes (Basel). 2021. PMID: 33919082 Free PMC article. Review.
-
DNA Methylation Clocks and Their Predictive Capacity for Aging Phenotypes and Healthspan.Neurosci Insights. 2020 Jul 21;15:2633105520942221. doi: 10.1177/2633105520942221. eCollection 2020. Neurosci Insights. 2020. PMID: 32743556 Free PMC article. Review.
-
Identification of Putative Markers That Predict the In Vitro Senescence of Mesenchymal Progenitor Cells.Cells. 2021 May 24;10(6):1301. doi: 10.3390/cells10061301. Cells. 2021. PMID: 34073789 Free PMC article.
-
Generation and Applications of Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells.Stem Cells Int. 2018 Jul 31;2018:9601623. doi: 10.1155/2018/9601623. eCollection 2018. Stem Cells Int. 2018. PMID: 30154868 Free PMC article. Review.
-
Embryonic Stem Cell-Derived Mesenchymal Stem Cells (MSCs) Have a Superior Neuroprotective Capacity Over Fetal MSCs in the Hypoxic-Ischemic Mouse Brain.Stem Cells Transl Med. 2018 May;7(5):439-449. doi: 10.1002/sctm.17-0260. Epub 2018 Feb 28. Stem Cells Transl Med. 2018. PMID: 29489062 Free PMC article.
References
-
- de Peppo G.M., Svensson S., Lennerås M., Synnergren J., Stenberg J., Strehl R., Hyllner J., Thomsen P., Karlsson C. Human embryonic mesodermal progenitors highly resemble human mesenchymal stem cells and display high potential for tissue engineering applications. Tissue Eng. Part A. 2010;16:2161–2182. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

