The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy
- PMID: 25220415
- PMCID: PMC4362780
- DOI: 10.1038/onc.2014.295
The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy
Abstract
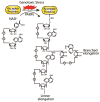
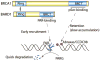
DNA damage is a deleterious threat, but occurs daily in all types of cells. In response to DNA damage, poly(ADP-ribosyl)ation, a unique post-translational modification, is immediately catalyzed by poly(ADP-ribose) polymerases (PARPs) at DNA lesions, which facilitates DNA damage repair. Recent studies suggest that poly(ADP-ribosyl)ation is one of the first steps of cellular DNA damage response and governs early DNA damage response pathways. Suppression of DNA damage-induced poly(ADP-ribosyl)ation by PARP inhibitors impairs early DNA damage response events. Moreover, PARP inhibitors are emerging as anti-cancer drugs in phase III clinical trials for BRCA-deficient tumors. In this review, we discuss recent findings on poly(ADP-ribosyl)ation in DNA damage response as well as the molecular mechanism by which PARP inhibitors selectively kill tumor cells with BRCA mutations.
Figures



Similar articles
-
Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions.Biochem J. 1999 Sep 1;342 ( Pt 2)(Pt 2):249-68. Biochem J. 1999. PMID: 10455009 Free PMC article. Review.
-
Poly(ADP-ribosyl)ation as a DNA damage-induced post-translational modification regulating poly(ADP-ribose) polymerase-1-topoisomerase I interaction.J Biol Chem. 2004 Sep 17;279(38):39686-96. doi: 10.1074/jbc.M402729200. Epub 2004 Jul 7. J Biol Chem. 2004. PMID: 15247263
-
Poly(ADP-ribosyl)ation in mammalian ageing.Nucleic Acids Res. 2007;35(22):7456-65. doi: 10.1093/nar/gkm735. Epub 2007 Oct 2. Nucleic Acids Res. 2007. PMID: 17913748 Free PMC article. Review.
-
Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function.Mol Cell Biol. 2003 Apr;23(7):2451-62. doi: 10.1128/MCB.23.7.2451-2462.2003. Mol Cell Biol. 2003. PMID: 12640128 Free PMC article.
-
Poly(ADP-ribosyl)ation, PARP, and aging.Sci Aging Knowledge Environ. 2004 Dec 8;2004(49):re9. doi: 10.1126/sageke.2004.49.re9. Sci Aging Knowledge Environ. 2004. PMID: 15590998 Review.
Cited by
-
Possible Novel Therapeutic Targets in Lymphangioleiomyomatosis Treatment.Front Med (Lausanne). 2020 Sep 24;7:554134. doi: 10.3389/fmed.2020.554134. eCollection 2020. Front Med (Lausanne). 2020. PMID: 33072782 Free PMC article. Review.
-
Increasing sensitivity to DNA damage is a potential driver for human ovarian cancer.Oncotarget. 2016 Aug 2;7(31):49710-49721. doi: 10.18632/oncotarget.10436. Oncotarget. 2016. PMID: 27391345 Free PMC article.
-
Bacterial Genotoxin-Induced DNA Damage and Modulation of the Host Immune Microenvironment.Toxins (Basel). 2020 Jan 21;12(2):63. doi: 10.3390/toxins12020063. Toxins (Basel). 2020. PMID: 31973033 Free PMC article. Review.
-
p21CDKN1A Regulates the Binding of Poly(ADP-Ribose) Polymerase-1 to DNA Repair Intermediates.PLoS One. 2016 Jan 5;11(1):e0146031. doi: 10.1371/journal.pone.0146031. eCollection 2016. PLoS One. 2016. PMID: 26730949 Free PMC article.
-
A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors.Cell Rep. 2015 Jun 9;11(9):1486-500. doi: 10.1016/j.celrep.2015.04.053. Epub 2015 May 21. Cell Rep. 2015. PMID: 26004182 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

