Short mitochondrial ARF triggers Parkin/PINK1-dependent mitophagy
- PMID: 25217637
- PMCID: PMC4207970
- DOI: 10.1074/jbc.M114.607150
Short mitochondrial ARF triggers Parkin/PINK1-dependent mitophagy
Abstract
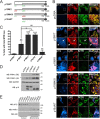
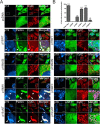
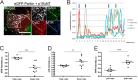
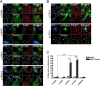
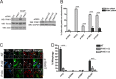
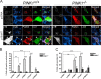
Parkinson disease (PD) is a complex neurodegenerative disease characterized by the loss of dopaminergic neurons in the substantia nigra. Multiple genes have been associated with PD, including Parkin and PINK1. Recent studies have established that the Parkin and PINK1 proteins function in a common mitochondrial quality control pathway, whereby disruption of the mitochondrial membrane potential leads to PINK1 stabilization at the mitochondrial outer surface. PINK1 accumulation leads to Parkin recruitment from the cytosol, which in turn promotes the degradation of the damaged mitochondria by autophagy (mitophagy). Most studies characterizing PINK1/Parkin mitophagy have relied on high concentrations of chemical uncouplers to trigger mitochondrial depolarization, a stimulus that has been difficult to adapt to neuronal systems and one unlikely to faithfully model the mitochondrial damage that occurs in PD. Here, we report that the short mitochondrial isoform of ARF (smARF), previously identified as an alternate translation product of the tumor suppressor p19ARF, depolarizes mitochondria and promotes mitophagy in a Parkin/PINK1-dependent manner, both in cell lines and in neurons. The work positions smARF upstream of PINK1 and Parkin and demonstrates that mitophagy can be triggered by intrinsic signaling cascades.
Keywords: Mitochondria; Mitophagy; PTEN-induced Putative Kinase 1 (PINK1); Parkin; Parkinson Disease; smARF.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review.
-
Nitric oxide induction of Parkin translocation in PTEN-induced putative kinase 1 (PINK1) deficiency: functional role of neuronal nitric oxide synthase during mitophagy.J Biol Chem. 2015 Apr 17;290(16):10325-35. doi: 10.1074/jbc.M114.624767. Epub 2015 Feb 25. J Biol Chem. 2015. PMID: 25716315 Free PMC article.
-
Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons.J Biol Chem. 2013 Jan 25;288(4):2223-37. doi: 10.1074/jbc.M112.391680. Epub 2012 Dec 4. J Biol Chem. 2013. PMID: 23212910 Free PMC article.
-
The Role of PTEN-L in Modulating PINK1-Parkin-Mediated Mitophagy.Neurotox Res. 2022 Aug;40(4):1103-1114. doi: 10.1007/s12640-022-00475-w. Epub 2022 Jun 14. Neurotox Res. 2022. PMID: 35699891 Review.
-
Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization.Hum Mol Genet. 2011 Mar 1;20(5):927-40. doi: 10.1093/hmg/ddq531. Epub 2010 Dec 7. Hum Mol Genet. 2011. PMID: 21147754 Free PMC article.
Cited by
-
Autophagy: Regulator of cell death.Cell Death Dis. 2023 Oct 4;14(10):648. doi: 10.1038/s41419-023-06154-8. Cell Death Dis. 2023. PMID: 37794028 Free PMC article. Review.
-
PINK1-PRKN mediated mitophagy: differences between in vitro and in vivo models.Autophagy. 2023 May;19(5):1396-1405. doi: 10.1080/15548627.2022.2139080. Epub 2022 Nov 3. Autophagy. 2023. PMID: 36282767 Free PMC article. Review.
-
miR-221-5p-Mediated Downregulation of JNK2 Aggravates Acute Lung Injury.Front Immunol. 2021 Nov 25;12:700933. doi: 10.3389/fimmu.2021.700933. eCollection 2021. Front Immunol. 2021. PMID: 34899681 Free PMC article.
-
Vorinostat in autophagic cell death: A critical insight into autophagy-mediated, -associated and -dependent cell death for cancer prevention.Drug Discov Today. 2022 Jan;27(1):269-279. doi: 10.1016/j.drudis.2021.08.004. Epub 2021 Aug 13. Drug Discov Today. 2022. PMID: 34400351 Free PMC article. Review.
-
Selective Neuron Vulnerability in Common and Rare Diseases-Mitochondria in the Focus.Front Mol Biosci. 2021 Jun 30;8:676187. doi: 10.3389/fmolb.2021.676187. eCollection 2021. Front Mol Biosci. 2021. PMID: 34295920 Free PMC article. Review.
References
-
- Corti O., Brice A. (2013) Mitochondrial quality control turns out to be the principal suspect in parkin and PINK1-related autosomal recessive Parkinson's disease. Curr. Opin. Neurobiol. 23, 100–108 - PubMed
-
- Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 - PubMed
-
- Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

