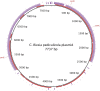
Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee lice
- PMID: 25213693
- PMCID: PMC4232544
- DOI: 10.1534/g3.114.012567
Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee lice
Abstract
The obligate-heritable endosymbionts of insects possess some of the smallest known bacterial genomes. This is likely due to loss of genomic material during symbiosis. The mode and rate of this erosion may change over evolutionary time: faster in newly formed associations and slower in long-established ones. The endosymbionts of human and anthropoid primate lice present a unique opportunity to study genome erosion in newly established (or young) symbionts. This is because we have a detailed phylogenetic history of these endosymbionts with divergence dates for closely related species. This allows for genome evolution to be studied in detail and rates of change to be estimated in a phylogenetic framework. Here, we sequenced the genome of the chimpanzee louse endosymbiont (Candidatus Riesia pediculischaeffi) and compared it with the closely related genome of the human body louse endosymbiont. From this comparison, we found evidence for recent genome erosion leading to gene loss in these endosymbionts. Although gene loss was detected, it was not significantly greater than in older endosymbionts from aphids and ants. Additionally, we searched for genes associated with B-vitamin synthesis in the two louse endosymbiont genomes because these endosymbionts are believed to synthesize essential B vitamins absent in the louse's diet. All of the expected genes were present, except those involved in thiamin synthesis. We failed to find genes encoding for proteins involved in the biosynthesis of thiamin or any complete exogenous means of salvaging thiamin, suggesting there is an undescribed mechanism for the salvage of thiamin. Finally, genes encoding for the pantothenate de novo biosynthesis pathway were located on a plasmid in both taxa along with a heat shock protein. Movement of these genes onto a plasmid may be functionally and evolutionarily significant, potentially increasing production and guarding against the deleterious effects of mutation. These data add to a growing resource of obligate endosymbiont genomes and to our understanding of the rate and mode of genome erosion in obligate animal-associated bacteria. Ultimately sequencing additional louse p-endosymbiont genomes will provide a model system for studying genome evolution in obligate host associated bacteria.
Keywords: Pediculus; gamma-proteobacteria; gene loss; genome erosion; primary-endosymbiont.
Copyright © 2014 Boyd et al.
Figures

Similar articles
-
Primates, Lice and Bacteria: Speciation and Genome Evolution in the Symbionts of Hominid Lice.Mol Biol Evol. 2017 Jul 1;34(7):1743-1757. doi: 10.1093/molbev/msx117. Mol Biol Evol. 2017. PMID: 28419279 Free PMC article.
-
Evolutionary relationships of "Candidatus Riesia spp.," endosymbiotic enterobacteriaceae living within hematophagous primate lice.Appl Environ Microbiol. 2007 Mar;73(5):1659-64. doi: 10.1128/AEM.01877-06. Epub 2007 Jan 12. Appl Environ Microbiol. 2007. PMID: 17220259 Free PMC article.
-
Two Bacterial Genera, Sodalis and Rickettsia, Associated with the Seal Louse Proechinophthirus fluctus (Phthiraptera: Anoplura).Appl Environ Microbiol. 2016 May 16;82(11):3185-97. doi: 10.1128/AEM.00282-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 26994086 Free PMC article.
-
Taxonomy of lice and their endosymbiotic bacteria in the post-genomic era.Clin Microbiol Infect. 2012 Apr;18(4):324-31. doi: 10.1111/j.1469-0691.2012.03782.x. Clin Microbiol Infect. 2012. PMID: 22429457 Review.
-
Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10255-61. doi: 10.1073/pnas.1423305112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26039986 Free PMC article. Review.
Cited by
-
Grandeur Alliances: Symbiont Metabolic Integration and Obligate Arthropod Hematophagy.Trends Parasitol. 2016 Sep;32(9):739-749. doi: 10.1016/j.pt.2016.05.002. Epub 2016 May 25. Trends Parasitol. 2016. PMID: 27236581 Free PMC article. Review.
-
Lightella neohaematopini: A new lineage of highly reduced endosymbionts coevolving with chipmunk lice of the genus Neohaematopinus.Front Microbiol. 2022 Aug 1;13:900312. doi: 10.3389/fmicb.2022.900312. eCollection 2022. Front Microbiol. 2022. PMID: 35979496 Free PMC article.
-
Endosymbiotic bacteria of the boar louse Haematopinus apri (Insecta: Phthiraptera: Anoplura).Front Microbiol. 2022 Aug 8;13:962252. doi: 10.3389/fmicb.2022.962252. eCollection 2022. Front Microbiol. 2022. PMID: 36003934 Free PMC article.
-
Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria.Nat Commun. 2024 May 29;15(1):4571. doi: 10.1038/s41467-024-48784-2. Nat Commun. 2024. PMID: 38811551 Free PMC article.
-
Legionella Becoming a Mutualist: Adaptive Processes Shaping the Genome of Symbiont in the Louse Polyplax serrata.Genome Biol Evol. 2017 Nov 1;9(11):2946-2957. doi: 10.1093/gbe/evx217. Genome Biol Evol. 2017. PMID: 29069349 Free PMC article.
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
