Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276
- PMID: 25203353
- PMCID: PMC4702256
- DOI: 10.1016/j.virol.2014.08.007
Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276
Abstract
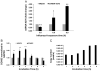
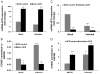
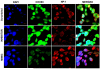
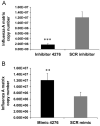
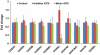
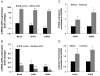
Influenza virus infection induces several changes in host miRNA profile, host cell death and tissue damage. Cytochrome c is a regulator of the intrinsic apoptotic pathway and is altered during viral infections. Within the first 3h of infection with influenza virus, significant down-regulation of hsa-miRNA-4276 (miRNA-4276) is followed by a 2-fold increase in cytochrome c oxidase VIC (COX6C) mRNA was found to occur in human alveolar and bronchial epithelial cells. Expression of caspase-9 also increased within the first 3h of infection, but subsequently decreased. Modulation of miR-4276 using mimic and inhibitor oligonucleotides showed significant down-regulation or up-regulation, respectively, of COX6C expression. Our data suggests that on initial exposure to influenza virus, host cells upregulate COX6C mRNA expression through silencing miR-4276 and repressed viral replication by inducing the apoptotic protein caspase-9. Taken together, these data suggest that miR-4276 may be an important regulator of the early stages of infection by influenza.
Keywords: COX6C; Influenza virus; Lung epithelial cells; miRNA.
Published by Elsevier Inc.
Figures

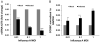

Similar articles
-
Upregulation of miRNA-4776 in Influenza Virus Infected Bronchial Epithelial Cells Is Associated with Downregulation of NFKBIB and Increased Viral Survival.Viruses. 2017 Apr 27;9(5):94. doi: 10.3390/v9050094. Viruses. 2017. PMID: 28448456 Free PMC article.
-
Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a.J Gen Virol. 2013 May;94(Pt 5):985-995. doi: 10.1099/vir.0.049528-0. Epub 2013 Jan 23. J Gen Virol. 2013. PMID: 23343627
-
Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway.Viruses. 2021 May 23;13(6):967. doi: 10.3390/v13060967. Viruses. 2021. PMID: 34071096 Free PMC article.
-
Cellular microRNAs influence replication of H3N2 canine influenza virus in infected cells.Vet Microbiol. 2021 Jun;257:109083. doi: 10.1016/j.vetmic.2021.109083. Epub 2021 Apr 20. Vet Microbiol. 2021. PMID: 33894663
-
Novel role of COX6c in the regulation of oxidative phosphorylation and diseases.Cell Death Discov. 2022 Jul 25;8(1):336. doi: 10.1038/s41420-022-01130-1. Cell Death Discov. 2022. PMID: 35879322 Free PMC article. Review.
Cited by
-
Herpes Simplex Virus 1 Suppresses the Function of Lung Dendritic Cells via Caveolin-1.Clin Vaccine Immunol. 2015 Aug;22(8):883-95. doi: 10.1128/CVI.00170-15. Epub 2015 May 27. Clin Vaccine Immunol. 2015. PMID: 26018534 Free PMC article.
-
High-Throughput MicroRNA Profiles of Permissive Madin-Darby Canine Kidney Cell Line Infected with Influenza B Viruses.Viruses. 2019 Oct 25;11(11):986. doi: 10.3390/v11110986. Viruses. 2019. PMID: 31717720 Free PMC article.
-
Novel Role for miR-1290 in Host Species Specificity of Influenza A Virus.Mol Ther Nucleic Acids. 2019 Sep 6;17:10-23. doi: 10.1016/j.omtn.2019.04.028. Epub 2019 May 15. Mol Ther Nucleic Acids. 2019. PMID: 31173947 Free PMC article.
-
Role of miRNA in Highly Pathogenic H5 Avian Influenza Virus Infection: An Emphasis on Cellular and Chicken Models.Viruses. 2024 Jul 9;16(7):1102. doi: 10.3390/v16071102. Viruses. 2024. PMID: 39066264 Free PMC article. Review.
-
Differential expression and clinical significance of COX6C in human diseases.Am J Transl Res. 2021 Jan 15;13(1):1-10. eCollection 2021. Am J Transl Res. 2021. PMID: 33527004 Free PMC article. Review.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Blachere FM, Cao G, Lindsley WG, Noti JD, Beezhold DH. Enhanced detection of infectious airborne influenza virus. J. Virol. Methods. 2011;176:120–124. - PubMed
-
- Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, Meade BJ, Lander O, Davis S, Thewlis RE, Celik I, Chen BT, Beezhold DH. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Diseases: an Official Publication Infect. Dis. Soc Am. 2009;48:438–440. - PubMed
-
- Brydon EW, Morris SJ, Sweet C. Role of apoptosis and cytokines in influenza virus morbidity. FEMS Microbiol. Rev. 2005;29:837–850. - PubMed
-
- Collins M. Potential roles of apoptosis in viral pathogenesis. Am. J. Respir. Crit. Care Med. 1995;152:S20–S24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

